Abstract
The LIM homeobox 2 (Lhx2) transcription factor Lhx2 has a variety of functions, including neural induction, morphogenesis, and hematopoiesis. Here we show the involvement of Lhx2 in osteoclast differentiation. Lhx2 was strongly expressed in osteoclast precursor cells but its expression was significantly reduced during receptor activator of nuclear factor-κB ligand (RANKL)-mediated osteoclastogenesis. Overexpression of Lhx2 in bone marrow-derived monocyte/macrophage lineage cells (BMMs), which are osteoclast precursor cells, attenuated RANKL-induced osteoclast differentiation by inhibiting the induction of nuclear factor of activated T cells c1 (NFATc1). Interestingly, interaction of Lhx2 proteins with c-Fos attenuated the DNA-binding ability of c-Fos and thereby inhibited the transactivation of NFATc1. Furthermore, Lhx2 conditional knockout mice exhibited an osteoporotic bone phenotype, which was related with increased osteoclast formation in vivo. Taken together, our results suggest that Lhx2 acts as a negative regulator of osteoclast formation in vitro and in vivo. The anti-osteoclastogenic effect of Lhx2 may be useful for developing a therapeutic strategy for bone disease.
Similar content being viewed by others
Main
Bone homeostasis is maintained by a delicate balance between the activities of osteoblasts and those of osteoclasts. Excessive osteoclast activity relative to osteoblast activity leads to bone diseases such as osteoporosis.1 Osteoblasts are mononuclear cells that are responsible for bone formation and osteoclast differentiation. Osteoblasts differentiate from mesenchymal progenitors under the control of various growth factors, in particular, the bone morphogenetic proteins (BMPs).2
Osteoclasts are unique bone cells that can control the amount of bone tissue by resorbing mineralized bone matrix. Osteoclasts are characterized by tartrate-resistant acid phosphatase (TRAP)-positive multinuclear cells (MNCs) originate from hematopoietic precursors. Osteoclast formation requires two essential cytokines: macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor κB ligand (RANKL). These membrane-bound proteins are produced by stromal cells and osteoblasts. Binding of M-CSF to its transmembrane receptor c-Fms induces receptor activation of nuclear κB (RANK) expression on osteoclast precursor cells by activating the Ets transcription factor PU.1 and thereby mediates the proliferation and survival of osteoclasts.3 Binding of RANKL to its receptor RANK activates various transcription factors, including c-Fos, PU.1, and nuclear factor of activated T cells c1 (NFATc1), which have a critical role in osteoclastogenesis.
c-Fos, a member of the AP-1 family of transcription factors, is induced by RANKL in the early stage of osteoclast differentiation. c-Fos knockout mice exhibit an osteopetrotic bone phenotype because of defects in the commitment of the osteoclast lineage.4, 5 Overexpression of c-Fos or NFATc1 in osteoclast precursor cells lacking c-Fos rescues the osteoclast differentiation defect.6 RANKL activates c-Fos, NF-κB, and a sustained low-amplitude calcium oscillation, which results in the induction and activation of NFATc1. Subsequently, activated NFATc1 induces the expression of target genes, including TRAP, cathepsin K, and osteoclast-associated receptor (OSCAR), which are important for osteoclast differentiation or function.4, 7, 8 NFATc1-deficient embryonic stem cells fail to differentiate into osteoclasts in response to RANKL.4 In addition, osteoclast-specific conditional NFATc1-deficient mice, which were generated to overcome the embryonic lethality of NFATc1 knockout mice, develop osteopetrosis as a result of the defect in osteoclastogenesis.9 Therefore, it has been well established that the RANKL/c-Fos/NFATc1 signaling pathway is the critical converging pathway for osteoclast differentiation via induction of various osteoclast marker genes such as TRAP, cathepsin K, and OSCAR.
The LIM homeobox 2 transcription factor Lhx2 (also known as LH2 or LH2A) is a member of the LIM homeodomain family of proteins. Members of the LIM-homeodomain family are made of a homeodomain and two LIM domains that comprise two zinc-finger-like structures. The LIM domain can associate with various transcriptional cofactors and may modulate the biological functions of LIM-homeodomain proteins.10 Lhx2 was originally identified in pituitary cells and in pre-B-cell lines.11, 12 Lhx2 expression is essential in a wide variety of progenitor/stem cell populations.13 Recent studies have shown that Lhx2 has important roles in eye development, liver differentiation, hair formation, and the complete development of olfactory sensory neurons.14, 15, 16
In this study, we conducted a microarray analysis to find new regulatory molecules that may act as negative regulators during osteoclastogenesis. As a result of the microarray analysis, we found that Lhx2 expression is strongly downregulated during osteoclast differentiation. Overexpression of Lhx2 in bone marrow-derived monocyte/macrophage lineage cells (BMMs) inhibits RANKL-mediated osteoclastogenesis by inhibiting binding of c-Fos to the promoter of NFATc1, which suggests that Lhx2 acts as a negative regulatory molecule during RANKL-mediated osteoclastogenesis. Furthermore, our results from osteoclast-specific deletion of Lhx2 clearly established that Lhx2 can control bone mass by regulating osteoclast differentiation in vivo.
Results
Lhx2 expression is downregulated by RANKL during osteoclast differentiation
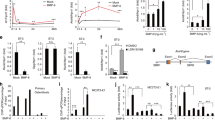
To examine the expression patterns of Lhx2, we performed real-time PCR in osteoclasts and osteoblasts. Osteoclast differentiation was induced by culturing BMMs in the presence of M-CSF and RANKL. Lhx2 expression was strongly downregulated by RANKL during osteoclast differentiation, whereas the expression of NFATc1 and TRAP, markers for osteoclasts, increased as the cells differentiated (Figure 1a). Because Lhx2 is downregulated by RANKL at the early stage of osteoclastogenesis, we determined which signaling pathways are essential for Lhx2 downregulation by using various inhibitors of early signaling pathways activated by RANKL. As shown in Supplementary Figure 1, treatment with SB203580 (p38 MAPK inhibitor), PD98059 (MEK inhibitor), and SP600125 (c-Jun N-terminal kinase (JNK) inhibitor) in BMMs did not affect Lhx2 expression downregulated by RANKL, whereas treatment with LY294002 (PI3K inhibitor) and TPCK (NF-κB inhibitor) significantly inhibited RANKL-induced Lhx2 downregulation. These results suggest that RANKL inhibits Lhx2 expression through AKT and NF-κB signaling pathways. To investigate the effect of pro-resorptive and anti-resorptive cytokines on Lhx2 expression, BMMs were treated with each cytokine overnight in the presence of M-CSF. Tumor necrosis factor (TNF)-α strongly reduced Lhx2 expression, whereas interferon (IFN)-α increased Lhx2 expression (Supplementary Figure 2). Other cytokines (interleukin (IL)-6, IFN-β, IFN-γ, and IL-4) showed effects in the opposite direction on Lhx2 expression.
Expression of Lhx2 in osteoclasts and osteoblasts. (a) BMMs were cultured with M-CSF and RANKL for the indicated times. (b) Osteoblasts were cultured with osteogenic medium containing BMP2, ascorbic acid, and β-glycerophosphate for the indicated times. (a and b) Quantitative real-time PCR was performed for mRNA expression of NFATc1, TRAP, Lhx2, ALP, and bone sialoprotein (BSP). Data represent means±S.Ds. *P<0.01, **P<0.001 versus control
Next, we examined the expression pattern of Lhx2 in primary osteoblasts cultured with osteogenic medium containing BMP2, ascorbic acid, and β-glycerophosphate. The expression of osteoblast marker genes including alkaline phosphatase (ALP) and bone sialoprotein was strongly increased during osteoblast differentiation, whereas Lhx2 expression was slightly increased (Figure 1b).
Overexpression of Lhx2 in osteoclast precursor cells inhibits RANKL-induced osteoclast formation
To investigate the role of Lhx2 in osteoclasts and osteoblasts, we overexpressed Lhx2 in BMMs and primary calvarial cells by using retrovirus. Transduced BMMs were cultured with M-CSF alone or M-CSF and RANKL and stained for TRAP. RANKL treatment of control vector-infected BMMs increased the number of TRAP(+) MNCs in a dose-dependent manner, whereas overexpression of Lhx2 in BMMs significantly inhibited the formation of TRAP(+) MNCs mediated by M-CSF and RANKL (Figures 2a and b). Next, to investigate the role of Lhx2 in osteoblasts, we examined the effect of Lhx2 on ALP activity and nodule formation in primary calvarial cells. Compared with osteoclasts, overexpression of Lhx2 had little effect on BMP-induced ALP activity and nodule formation (Figures 2c and d). Taken together, these data suggested that Lhx2 has a critical role in RANKL-mediated osteoclast differentiation.
Overexpression of Lhx2 in BMMs attenuates RANKL-induced osteoclast differentiation. (a and b) BMMs were infected with pMX-IRES-EGFP (control) or Lhx2 retrovirus. Transduced BMMs were cultured with M-CSF alone or M-CSF and RANKL as indicated for 3 days. (a) Cells were fixed and stained for TRAP. (b) Numbers of TRAP-positive multinucleated cells per well were counted. Data represent means±S.Ds. *P<0.01 versus control. (c and d) Osteoblasts were infected with pMX-IRES-EGFP (control) or Lhx2 retrovirus. Transduced osteoblasts were cultured without or with osteogenic medium containing BMP2, ascorbic acid, and β-glycerophosphate. (c) Cells were cultured for 4 days and ALP activities were measured by densitometry at 405 nm. Data represent means±S.Ds. (d) Cells cultured for 9 days were fixed and stained for alizarin red
1,25-Dihydroxyvitamin D3 (1,25(OH)2D3) indirectly stimulates osteoclast differentiation by increasing RANKL and decreasing osteoprotegerin (OPG) in osteoblasts.17 Thus, we investigated whether Lhx2 has a role in 1,25(OH)2D3-mediated osteoclast differentiation. Consistent with previous results,1, 18 the treatment of primary calvarial cells with 1,25(OH)2D3 induced RANKL expression and reduced OPG expression. However, it did not show any effect on Lhx2 expression (Supplementary Figure 3). In addition, when BMMs were cocultured with transduced primary osteoblasts in the presence of 1,25(OH)2D3 alone or 1,25(OH)2D3 and PGE2, the number of TRAP(+) MNCs was not changed by overexpression of Lhx2 in osteoblasts compared with the control (Supplementary Figures 4a and b). Therefore, these results suggested that Lhx2 may modulate osteoclast differentiation via direct action on osteoclast precursors rather than via indirect action on osteoblasts.
Overexpression of Lhx2 attenuates NFATc1 expression
Given our observation that overexpression of Lhx2 inhibits RANKL-mediated osteoclast differentiation, we examined the effect of Lhx2 on RANKL-induced early signaling pathways such as NF-κB, JNK, p38, and extracellular signal-regulated kinase (ERK). Among these signaling pathways, RANKL-induced activation of JNK, but not other signaling cascades, was slightly attenuated by overexpression of Lhx2 in BMMs (Supplementary Figure 5). Next, we investigated whether overexpression of Lhx2 could affect the expression of genes known to have a role in osteoclastogenesis. The expression of c-Fos at an early stage of osteoclastogenesis was not affected by overexpression of Lhx2 (Figures 3a and b). However, compared with control, exogenous overexpression of Lhx2 attenuated the expression of NFATc1 as well as of marker genes such as TRAP and OSCAR during RANKL-mediated osteoclastogenesis (Figures 3c and d). Therefore, these results further confirmed that Lhx2 has a role in osteoclast differentiation by modulating signals downstream from RANKL.
Overexpression of Lhx2 in BMMs inhibits expression of NFATc1 and OSCAR during RANKL-induced osteoclast differentiation. (a–d) BMMs were infected with pMX-IRES-EGFP (control) or Lhx2 retrovirus. Transduced BMMs were cultured with M-CSF alone or M-CSF and RANKL for the indicated times. (a and c) Quantitative real-time PCR was performed for the mRNA expression of Lhx2, c-Fos, NFATc1, TRAP, and OSCAR. Data represent means±S.Ds. *P<0.01, **P<0.001 versus control. (b and d) Cells were harvested from each time point and lysates were analyzed by western blot analysis with specific antibodies as indicated
Lhx2 attenuates the binding ability of c-Fos
Because overexpression of Lhx2 attenuated RANKL-induced NFATc1 expression, we investigated whether Lhx2 could regulate the ability of c-Fos, an upstream transcription factor of NFATc1, to induce NFATc1 gene expression. We used a reporter assay involving transient transfection into highly transfectable 293 human embryonic kidney cells (293T). When a reporter plasmid containing a 6.2-kb NFATc1 promoter region was cotransfected with c-Fos, relative luciferase activity was increased (Figure 4a). However, Lhx2 decreased the induction of luciferase activity by c-Fos in a dose-dependent manner.
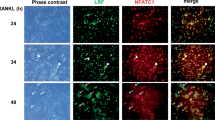
Lhx2 inhibits the transcriptional activity of c-Fos through direct interaction. (a) A NFATc1 6.2-kb promoter luciferase reporter was co-transfected with c-Fos and increasing amounts of Lhx2 into 293T cells. At 2 days after transfection, cells were lysed and luciferase activities were analyzed. Data represent means±S.Ds. **P<0.001 versus control. (b) 293T cells were transfected with Flag-tagged c-Fos plasmid. Cells were lysed and cell lysates were pulled down with glutathione S-transferase (GST) or GST–Lhx2 fusion proteins immobilized on glutathione-Sepharose beads. Pulled-down samples and whole-cell extracts (WCE) were subjected to SDS-PAGE and were analyzed by western blot analysis with specific antibodies as indicated. (c) c-Fos lysates were prepared by using TNT rabbit reticulocyte lysates. 32P-Labeled probe spanning the AP-1-binding sites in the mouse NFATc1 promoter was incubated with c-Fos lysates in the presence of GST or GST-Lhx2 proteins. To determine specific binding, an excess of cold, unlabeled wild-type or mutant probe was added to the mixture of labeled probe and c-Fos lysates. The protein/DNA complexes were separated on a nondenaturing polyacrylamide gel. (d) pMX-IRES-EGFP (control) or Lhx2 retrovirus-infected BMMs were cultured with M-CSF and RANKL for 1 day before cross-linking. Samples were immunoprecipitated with control IgG or anti-c-Fos antibody for chromatin immunoprecipitation (ChIP) analysis. PCR analysis was carried out with primers containing the AP1-binding sites in the NFATc1 promoter region
To investigate the inhibitory mechanism of Lhx2 on c-Fos transcription activity, we used a glutathione S-transferase (GST) pull-down assay to determine the interaction between Lhx2 and c-Fos. GST alone or GST-Lhx2 fusion proteins immobilized on glutathione-Sepharose beads were incubated with cell lysates containing Flag-tagged c-Fos. As shown in Figure 4b, c-Fos was retained by GST-Lhx2, but not by GST alone. Next, we also investigated whether endogenous Lhx2 and c-Fos interact with each other in preosteoclasts. We confirmed a direct interaction between Lhx2 and c-Fos in preosteoclasts (Supplementary Figure 6).
To determine whether Lhx2 proteins can modulate the binding of c-Fos to AP1-binding sites in the NFATc1 promoter region, we carried out an electrophoretic mobility shift assay (EMSA) in the presence of GST-Lhx2 fusion proteins. We observed a shift in the c-Fos-specific band resulting from the reaction mixture containing c-Fos translated in vitro with the probe containing the AP1-binding site. The specificity of this binding was confirmed by competition studies using cold wild-type and mutant competitor probes. When the purified GST-Lhx2 fusion proteins were added to the reaction mixture, we observed a significant decrease in c-Fos binding to labeled probe caused by GST-Lhx2 proteins, but not by GST alone (Figure 4c). Consistent with the EMSA data, chromatin immunoprecipitation (ChIP) assays showed that overexpression of Lhx2 in BMMs strongly attenuated an increase in c-Fos binding to the NFATc1 promoter region mediated by RANKL treatment compared with control (Figure 4d). Collectively, these results suggested that Lhx2 proteins decrease c-Fos binding to AP1-binding sites in the NFATc1 promoter region by association with c-Fos.
Downregulation of Lhx2 enhances osteoclastogenesis and expression of NFATc1 and target genes
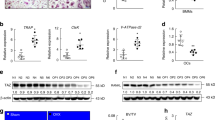
Because Lhx2 acts as a negative regulator of osteoclastogenesis, we investigated its physiological role in osteoclastogenesis by use of siRNAs. A net decrease in mRNA expression was observed in osteoclasts transfected with Lhx2-specific siRNA compared with control GFP siRNA (Figure 5c). siRNA-transfected BMMs were cultured for 4 days with M-CSF alone or with M-CSF and various concentrations of RANKL. RANKL treatment of control GFP siRNA-transfected BMMs increased the number of TRAP+ MNCs in a dose-dependent manner (Figures 5a and b). Compared with the control siRNA, the silencing of Lhx2 in BMMs resulted in a significant increase in the formation of TRAP+ MNCs mediated by RANKL.
Downregulation of Lhx2 by siRNAs enhances RANKL-induced osteoclast differentiation. (a and b) BMMs transfected with control or Lhx2 siRNAs were cultured with M-CSF and increasing concentrations of RANKL for 4 days. (a) Cells were fixed and stained for TRAP. (b) Numbers of TRAP-positive multinucleated cells per well were counted. Data represent means±S.Ds. #P<0.05, **P<0.001 versus control. (c) BMMs transfected with control or Lhx2 siRNAs were cultured with M-CSF and RANKL for 2 days. Quantitative real-time PCR was performed for the mRNA expression of Lhx2, NFATc1, TRAP, and OSCAR. Data represent means±S.Ds. #P<0.05, *P<0.01, **P<0.001 versus control
In addition, the silencing of Lhx2 in BMMs resulted in a significant increase in the induction of NFATc1 and marker genes such as TRAP and OSCAR in response to RANKL stimulation (Figure 5c). Taken together, these results suggested that Lhx2 has an important role in RANKL-induced osteoclastogenesis.
Deletion of Lhx2 results in reduced bone mass in vivo
To examine the effect of Lhx2 on the bone phenotype and osteoclast formation in vivo, we generated Mx1-cre-mediated Lhx2 conditional knockout mice. Control (Lhx2fl/fl) and Lhx2 conditional knockout mice (Lhx2fl/fl;Mx1-Cre) were intraperitoneally injected with polyinosinic-polycytidylic acid (polyI:C) every other day for a total of three doses at 2 weeks of age to induce Mx1-cre expression. We confirmed the reduction of Lhx2 expression in long bone, osteoclast precursor cells, and osteoclasts of Lhx2 conditional knockout mice by using quantitative real-time PCR analysis (Figures 6c–e). Consistent with results from experiments using siRNAs, in BMMs derived from Lhx2 conditional knockout mice, RANKL-induced osteoclast differentiation was significantly increased (Figures 6a and b). Also, Mx1-cre-mediated reduction of Lhx2 in BMMs resulted in a significant increase in the induction of NFATc1, TRAP, and OSCAR mediated by RANKL (Figure 6e).
BMMs derived from Lhx2 conditional knockout mice result in enhanced osteoclast differentiation mediated by RANKL. (a and b) BMMs derived from control or Lhx2 conditional knockout mice were cultured with M-CSF and increasing concentrations of RANKL for 3 days. (a) Cultured cells were fixed and stained for TRAP. (b) Numbers of TRAP-positive multinucleated cells per well were counted. Data represent means±S.Ds. **P<0.001 versus control. (c) Total RNA was harvested from the long bone of control or Lhx2 conditional knockout mice and mRNA expression of Lhx2 was assessed by quantitative real-time PCR. Data represent means±S.Ds. *P<0.01 versus control. (d) BMMs derived from control or Lhx2 conditional knockout mice were cultured with M-CSF alone or M-CSF and RANKL for 2 days. mRNA expression of Lhx2 was assessed by quantitative real-time PCR. Data represent means±S.Ds. #P<0.05 versus control. (e) BMMs derived from control or Lhx2 conditional knockout mice were cultured with M-CSF and RANKL for 2 days and quantitative real-time PCR was performed for the mRNA expression of Lhx2, NFATc1, TRAP, and OSCAR. Data represent means±SDs. *P<0.01, **P<0.001 versus control
Next, we analyzed the bone phenotype to examine the effect of Lhx2 deletion on osteoclast formation in vivo. Micro-computed tomography (μCT) analysis showed a decrease in bone mass in Lhx2 conditional knockout mice compared with control mice (Figure 7a). The percentage of bone volume to total tissue volume and the trabecular number of Lhx2 conditional knockout mice were lower than those of control mice, which suggests that Lhx2 conditional knockout mice exhibit an osteoporotic phenotype (Figure 7b). To further confirm the phenotype of Lhx2 conditional knockout mice, we performed histomorphometric analysis in the proximal tibiae. As shown in Figures 7c–e, both osteoclast number and osteoclast surface area were significantly increased in Lhx2 conditional knockout mice (Figures 7c and e), whereas osteoblast number was not significantly different between control mice and Lhx2 conditional knockout mice (Figure 7e). Therefore, these results indicated that reduction of Lhx2 resulted in the phenotype of decreased bone mass with increased osteoclast formation in vivo.
Lhx2 conditional knockout mice exhibit an osteoporotic bone phenotype. (a) Representative three-dimensional reconstruction images of femurs in control or Lhx2 conditional knockout mice. (b) Bone parameters including bone volume per tissue volume (BV/TV), trabecular bone thickness (Tb.Th), trabecular separation (Tb.Sp), and trabecular number (Tb.N) were assessed by quantitative micro-CT. Data represent means±SDs (n=6). *P<0.01 versus control. (c) The TRAP staining of histological sections of proximal tibiae. (d) Hematoxylin/eosin (H&E) staining of histological sections of proximal tibiae. (e) Osteoclast surface per bone surface, osteoclast number per bone surface, and osteoblast number per bone surface were assessed. Data represent means±S.Ds. (n=3). #P<0.05, *P<0.01 versus control
Discussion
Homeobox genes have a major role in development.19 LIM-homeodomain proteins are encoded by a subfamily of homeobox-containing genes. The LIM domain recognizes a variety of transcriptional cofactors through protein–protein interaction. Thus, combinational interaction of LIM domains with other transcriptional regulators in a homometric or heterometric manner enables LIM-homeodomain proteins to participate in a wide range of developmental events by means of tissue-specific transcriptional regulation.10, 19
Here, for the first time, we suggest a novel role of Lhx2, a member of the LIM-homeodomain family of proteins, as a negative regulator of RANKL-induced osteoclast differentiation. We found that RANKL strongly downregulates Lhx2 expression during osteoclast differentiation. Overexpression of Lhx2 in BMMs attenuates RANKL-induced osteoclast differentiation by downregulation of NFATc1, which is a key modulator for osteoclast differentiation. In addition, we observed that Lhx2 interacts directly with c-Fos and inhibits its transcriptional activity by blocking recruitment of c-Fos to AP1-binding sites of the NFATc1 promoter. Thus, inhibition of Lhx2 expression by RANKL at the early stage of osteoclast differentiation likely enables c-Fos to bind to the NFATc1 promoter region. Subsequently, NFATc1 leads to the robust induction of TRAP and OSCAR, which are important for osteoclast differentiation and function. Therefore, Lhx2 is responsible for the negative regulation of RANKL-induced osteoclast differentiation by means of its ability to inhibit the transcriptional activity of c-Fos.
Previous studies have suggested that Lhx2 can act as a transcriptional activator or repressor by means of several mechanisms depending on the target genes. Lhx2 can activate transcription of the thyroid-stimulating hormone β subunit gene through direct binding to the Lhx2-binding region of the promoter.20 Hou et al. demonstrated that Lhx2 binds to enhancers rather than the promoter of PAX6 to induce neural differentiation from human embryonic stem cells.21 Also, the LIM domain of Lhx2 functions as a coactivator through interaction with MRG1, leading to recruitment of p300/CBP to stimulate glycoprotein hormone α-subunit gene expression.22 On the other hand, Lhx2 negatively regulates the PRL promoter in pituitary cell lines by an as yet unknown mechanism.23 In the present study, we elucidated the mechanism of action of Lhx2 as a negative regulator during osteoclastogenesis. The expression patterns of c-Fos, NFATc1, and Lhx2 during RANKL-induced osteoclastogenesis suggest that Lhx2 may repress c-Fos. During osteoclastogenesis, RANKL strongly induces the expression of c-Fos and NFATc1 and simultaneously downregulates the expression of Lhx2. In addition, overexpression of Lhx2 in BMMs attenuates transcription of the target gene of c-Fos. In our ChIP assays and EMSA assay, Lhx2 did not appear to directly bind to the NFATc1 promoter but appeared to inhibit binding of c-Fos to the promoter region of NFATc1. In addition, given our results that Lhx2 proteins interact with c-Fos in vitro, these findings suggest a model in which Lhx2 represses transcription of the target gene of c-Fos through interaction with this important transcription factor in osteoclasts.
Because Lhx2 knockout mice die a few days before birth owing to severe anemia, we generated Mx1-cre-mediated Lhx2 conditional knockout mice to confirm the effect of Lhx2 on the bone phenotype in vivo.16 Mx1-cre-mediated deletion of Lhx2 in mice resulted in an osteoporotic bone phenotype owing to increased osteoclast differentiation. Our Lhx2 conditional knockout mice exhibited no significant effect on osteoblast differentiation. These results may be caused from use of the inducible Mx1-cre and polyI:C system to specifically delete Lhx2 in IFN-responsive cells including monocyte and macrophage cells. However, our Lhx2 conditional knockout mice suggest that Lhx2 regulates bone remodeling in vivo by inhibiting osteoclast formation. Our results in the conditional knockout mice also suggest that the expression levels of Lhx2 in osteoclasts have no effect on osteoblast differentiation regulated by osteoclasts.
It is known that Lhx2 has an important role in regulating extracellular signaling pathways including the WNT and BMP pathways during neural development. Lhx2 activates antagonists of WNT and BMP protein in certain areas to inhibit these signaling pathways in the developing forebrain.21, 24 Because WNT and BMP signaling are involved in osteoblast differentiation and bone formation, we examined whether Lhx2 directly affects osteoblast differentiation and function.25 Overexpression of Lhx2 in osteoblasts showed a marginal effect on BMP2-mediated osteoblast differentiation and nodule formation (Figures 2c and d). Therefore, our and previous results collectively suggest that Lhx2 differentially regulates downstream signaling in a cell-type-specific manner. BMPs can function in osteoclasts as well as in osteoblasts. BMPs act synergistically with RANKL for induction of osteoclast formation from osteoclast precursors and directly stimulate mature osteoclast function mediated by downstream molecules expressed in osteoclasts.26 Although Lhx2 may have no effect on downstream molecules of BMP2 expressed in osteoblasts, we cannot be sure whether Lhx2 regulates the BMP signaling pathway during RANKL-induced osteoclastogenesis. Thus, further study will be required to elucidate the role of Lhx2 in osteoclast differentiation and function mediated by the BMP signaling pathway.
In our and previous results, it was shown that RANKL inhibits the expression of the MafB, Ids, and IRF8 genes, which are negative regulators of osteoclastogenesis.27, 28, 29 Here, we add Lhx2 to this list of negative regulators. Taken together, our results demonstrate that RANKL not only activates positive regulators such as c-Fos and NFATc1 but also represses negative regulators such as MafB, Ids, and Lhx2 to maximize the ability of the positive regulators for sufficient osteoclast formation.
TNF-α as well as RANKL represses the expression of Lhx2 (Supplementary Figure 2). TNF-α, a proinflammatory cytokine, may be responsible for osteoclast differentiation and activation in inflammatory bone diseases such as rheumatoid arthritis.30, 31 Interestingly, we confirmed that overexpression of Lhx2 in BMMs significantly inhibited the formation of TRAP(+) MNCs mediated by M-CSF and TNF-α (Supplementary Figure 7). Therefore, further study of the precise mechanism of Lhx2 in osteoclasts may provide a new therapeutic strategy for bone diseases including osteoporosis and rheumatoid arthritis.
Materials and Methods
Reagents
Antibodies specific for Flag and actin were purchased from Sigma-Aldrich (St. Louis, MO, USA); c-Fos and NFATc1 were from Santa Cruz Biotechnology (Santa Cruz, CA, USA); and GST, phospho-p38, p38, phospho-JNK, JNK, phospho-Erk, Erk, and IkB were from Cell Signaling Technology (Beverly, MA, USA). A polyclonal antibody for OSCAR was prepared as previously described.7 TNF-α, IL-1, IL-6, and IL-4 were purchased from R&D systems (Minneapolis, MN, USA).
Osteoclast differentiation
Mouse bone marrow cells were isolated from tibiae and femurs of 6-week-old mice by flushing the bone marrow with α-minimal essential medium (α-MEM). Bone marrow cells were cultured in α-MEM containing 10% fetal bovine serum with M-CSF (30 ng/ml) for 3 days. Adherent cells were used as osteoclast precursors (BMMs) and BMMs were further cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 3 days. Primary osteoblasts and BMMs were cocultured for 5 days in the presence of 1,25(OH)2D3 (1 × 10−8 M) and PGE2 (1 × 10−6 M). Cultured cells were fixed and stained with TRAP solution (Sigma-Aldrich) according to the manufacturer’s instructions. TRAP-positive cells with more than three nuclei were counted as TRAP-positive MNCs. Cells were observed by using a Leica DMIRB microscope equipped with an N Plan 10 × 0.25 numerical aperture objective lens (Leica, Wetzlar, Germany). Images were captured by using a ProgRes Capture Pro (Jenoptik, Jena, Germany).
Osteoblast differentiation
Primary osteoblasts were prepared from calvariae of newborn mice by enzymatic digestion in α-MEM with 0.1% collagenase (Invitrogen, Carlsbad, CA, USA) and 0.2% dispase (Roche Applied Sciences, Indianapolis, IN, USA). To induce osteoblast differentiation, cells were cultured in α-MEM containing 10% fetal bovine serum with BMP2 (50 ng/ml), β-glycerophosphate (100 mM), and ascorbic acid (50 μg/ml). For the ALP activity assay, cells were lysed at day 4 of culture and cell lysates were incubated with the enzyme’s nonspecific substrate, p-nitrophenyl phosphate (Sigma-Aldrich). ALP activity was measured spectrophotometrically at 405 nm. For alizarin red staining, cells were fixed at day 9 of culture and stained with 40 mM pH 4.2 alizarin red solution.
Retroviral infection
To prepare retroviral supernatants, retroviral vectors were transfected into the packaging cell line Plat E by using FuGENE 6 (Roche Applied Sciences) according to the manufacturer’s protocol. Viral supernatants were collected from cultured media 48 h after transfection. BMMs or osteoblasts were incubated with viral supernatants for 8 h in the presence of Polybrene (10 μg/ml).
Real-time PCR
Total RNA was extracted from cultured cells by using Qiazol lysis reagent (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. An amount of 2 μg of RNA was reverse transcribed into cDNA by using Superscript II Reverse Transcriptase (Invitrogen). Quantitative real-time PCR analysis was performed in triplicate with a Rotor-Gene Q (Qiagen) using SYBR Green (Qiagen). The thermal cycling conditions were as follows: 15 min at 95 °C, followed by 40 cycles of 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. Amounts of mRNAs were normalized to an endogenous control glyceraldehyde-3-phosphate dehydrogenase (Gapdh). The relative quantitation value for each target gene compared with the calibrator for that target was expressed as 2−(Ct-Cc) (Ct and Cc are the mean threshold cycle differences after normalizing to Gapdh). The relative expression levels of samples were presented by semi-log plot. The primer sequences are as follows: c-Fos, 5′-ATGGGCTCTCCTGTCAACACA-3′ and 5′-TGGCAATCTCAGTCTGCAACGCAG-3′; OSCAR, 5′-TGCTGGTAACGGATCAGCTCCCCAGA-3′ and 5′-CCAAGGAGCCAGAACCTTCGAAACT-3′; Alp, 5′-CAAGGATATCGACGTGATCAT G-3′ and 5′-GTCAGTCAGGTTGTTCCGATT C-3′; bone sialoprotein, 5′-GGAAGAGGAGACTTCAAACGAAG-3′ and 5′-CATCCACTTCTGCTTCTTCGTTC-3′; Lhx2, 5′-GCAAAAGACCGGCCTCACCAAGAG-3′ and 5′-AAAGGTTGGTAAGAGTCGTTTGTG-3′; NFATc1, 5′-CTCGAAAGACAGCACTGGAGCAT-3′ and 5′’-CGGCTGCCTTCCGTCTCATAG-3′; TRAP, 5′-CTGGAGTGCACGATGCCAGCGACA-3′ and 5′-TCCGTGCTCGGCGATGGACCAGA-3′; RANKL, 5′-CCTGAGACTCCATGAAAACGC-3′ and 5′-TCGCTGGGCCACATCCAACCATGA-3′; OPG, 5′-CGGCGTGGTGCAAGCTGGAAC-3′ and 5′-CCTCTTCACACAGGGTGACATC-3′; GAPDH, 5′-TGACCACAGTCCATGCCATCACTG-3′ and 5′-CAGGAGACAACCTGGTCCTCAGTG-3′.
Luciferase assay
293T cells were plated on 24-well plates at a density of 2 × 104 cells per well 1 day before transfection. Reporter plasmid and various expression vectors were cotransfected into the cells by using FuGENE 6 according to the manufacturer’s instruction. After 2 days of transfection, the cells were lysed in reporter lysis buffer (Promega, Madison, WI, USA) and then luciferase activity was measured with a dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions.
GST pull-down assay
GST or GST-Lhx2 proteins were expressed in Escherichia coli BL21 (DE) and purified with glutathione-Sepharose 4B beads (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer’s instructions. 293T cells were transfected with Flag-c-Fos and lysed in 0.5% Nonidet P-40 lysis buffer. Cell lysates were incubated with either GST or GST-Lhx2 proteins for 3 h and further incubated with glutathione-Sepharose 4B beads. Precipitated samples were subjected to SDS-PAGE and western blotting.
EMSA
c-Fos proteins were synthesized by using a TNT-coupled rabbit reticulocyte lysate system (Promega). An amount of 4 μg of TNT cell lysate, 2 μg of poly (dI-dC), and 50 000 cpm-labeled probes were incubated in reaction buffer (10 mM HEPE, pH 8.0, 0.5 mM dithiothreitol, 50 mM KCL, 5 mM MgCl2, 1 mM EDTA, protease inhibitors, and 5% glycerol) at 25 °C for 30 min. Samples were resolved on a 5% native polyacrylamide gel in 0.25 TBE buffer. Wild- and mutant-type sequences of AP1-binding sites were as follows: wild-type AP1-binding site, 5′-AGCCCGGCCCTGCGTCAGAGTGAGAC-3′ and 5′-GTCTCACTCTGACGCAGGGCCGGGCT-3′, mutant-type AP1-binding site, 5′-AGCCCGGCCCAACGTAGGAGTGAGAC-3′ and 5′-GTCTCACTCCTACGTTGGGCCGGGCT-3′.
ChIP assay
A ChIP assay was performed with the ChIP kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Briefly, BMMs were transduced with pMX-IRES-EGFP (control) or Lhx2 retrovirus and were cultured with M-CSF and RANKL for 1 day. Cultured cells were cross-linked with 1% formaldehyde for 10 min at 37 °C and were lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1, 0.01% protease inhibitor mixture). Cells were sonicated to shear the DNA into small fragments of an average length between 200 and 1000 base pairs. Diluted supernatants were immunoprecipitated with anti-c-Fos or control IgG (SantaCruz Biotechnology), washed, and then eluted in fresh elution buffer (0.1 M NaHCO3 and 1% SDS). After cross-link reversal and Proteinase K treatment, DNA samples were recovered by phenol/chloroform extraction and ethanol precipitation. The purified DNA samples were subjected to PCR amplification with primers specific for the promoter regions of NFATc1-containing c-Fos-binding sites. The primer sequences are as follows; NFATc1, 5′-CCGGGACGCCCATGCAATCTGTTAGTAATT-3, and 5′-GCGGGTGCCCTGAGAAAGCTACTCTCCCTT-3′.
siRNA transfection
Control siRNAs and Lhx2 siRNAs were purchased from Dharmacon (Lafayette, CO, USA). The siRNAs were transfected into BMMs by using lipofectamine 2000 (Invitrogen).
Lhx2 conditional knockout mice
Lhx2fl/fl mice were previously generated and have been described.32 To generate Lhx2 conditional knockout mice (Lhx2fl/fl;Mx1-Cre), Lhx2fl/fl mice were crossed with Mx1-Cre mice expressing Cre recombinase under the control of a type I IFN-inducible Mx1 promoter. Lhx2fl/+;Mx1-Cre mice were crossed with Lhx2fl/fl mice and then Lhx2fl/fl mice were crossed with Lhx2fl/fl;Mx1-Cre mice. Control (Lhx2fl/fl) and Lhx2 conditional knockout mice (Lhx2fl/fl;Mx1-Cre) at 2 weeks of age were intraperitoneally injected with 200 μg of polyI:C every other day for a total of three doses to activate Mx1-Cre.
μCT and histomorphometric analyses
We analyzed the femurs of 10-week-old female mice by using μCT (Skyscan 1172, Skyscan, Kontich, Belgium). The X-ray source was set at 50 kV and 200 A with a pixel size of 17.09 mm. Exposure time was 1.2 s. Four-hundred and fifty projections were acquired over an angular range of 180° (angular step of 0.4°). The image slices were reconstructed by using 3D CT analyzer software (CTAN, Skyscan). Trabecular morphometry was characterized by measuring the bone volume fraction and trabecular thickness. For static histomorphometry, the tibia from each mouse was removed and fixed in 4% paraformaldehyde in PBS at 4 °C, decalcified in 12% EDTA for 4 weeks, and embedded in paraffin. Tissue sections of 5-μm thickness were subjected to TRAP staining or hematoxylin/eosin staining according to standard procedures. Histomorphometric parameters were calculated on the basis of standard formulas, as described previously.33
Abbreviations
- RANKL:
-
receptor activator of nuclear factor-κB ligand
- 1,25(OH)2D3:
-
1α,25-dihydroxyvitamin D3
- BMP:
-
bone morphogenetic protein
- ALP:
-
alkaline phosphatase
- TRAP:
-
tartrate-resistant acid phosphate
- M-CSF:
-
macrophage colony-stimulating factor
- JNK:
-
c-Jun N-terminal kinase
- ERK:
-
extracellular signal-regulated kinase
- NFAT:
-
nuclear factor of activated T cells
- TNF:
-
tumor necrosis factor
- IFN:
-
interferon
- IL-6:
-
interleukin-6
- OSCAR:
-
osteoclast-associated receptor
- OPG:
-
osteoprotegerin
- BMM:
-
bone marrow-derived macrophage lineage cell
- ChIP:
-
chromatin immunoprecipitation
- Lhx2:
-
LIM homeobox 2
- NFATc1:
-
nuclear factor of activated T cells c1
- MNC:
-
multinuclear cell
- EMSA:
-
electrophoretic mobility shift assay
References
Walsh MC, Kim N, Kadono Y, Rho J, Lee SY, Lorenzo J et al. Osteoimmunology: interplay between the immune system and bone metabolism. Annu Rev Immunol 2006; 24: 33–63.
Agata H, Asahina I, Yamazaki Y, Uchida M, Shinohara Y, Honda MJ et al. Effective bone engineering with periosteum-derived cells. J Dent Res 2007; 86: 79–83.
Arai F, Miyamoto T, Ohneda O, Inada T, Sudo T, Brasel K et al. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. J Exp Med 1999; 190: 1741–1754.
Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 2002; 3: 889–901.
Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA et al. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science 1994; 266: 443–448.
Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZ et al. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem 2004; 279: 26475–26480.
Kim K, Kim JH, Lee J, Jin HM, Lee SH, Fisher DE et al. Nuclear factor of activated T cells c1 induces osteoclast-associated receptor gene expression during tumor necrosis factor-related activation-induced cytokine-mediated osteoclastogenesis. J Biol Chem 2005; 280: 35209–35216.
Matsumoto M, Kogawa M, Wada S, Takayanagi H, Tsujimoto M, Katayama S et al. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J Biol Chem 2004; 279: 45969–45979.
Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM et al. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest 2008; 118: 3775–3789.
Dawid IB, Breen JJ, Toyama R . LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet 1998; 14: 156–162.
Bach I, Rhodes SJ, Pearse RV 2nd, Heinzel T, Gloss B, Scully KM et al. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA 1995; 92: 2720–2724.
Xu Y, Baldassare M, Fisher P, Rathbun G, Oltz EM, Yancopoulos GD et al. LH-2: a LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc Natl Acad Sci USA 1993; 90: 227–231.
Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 1997; 124: 2935–2944.
Tetreault N, Champagne MP, Bernier G . The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol 2009; 327: 541–550.
Tornqvist G, Sandberg A, Hagglund AC, Carlsson L . Cyclic expression of lhx2 regulates hair formation. PLoS Genet 2010; 6: e1000904.
Hirota J, Mombaerts P . The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci USA 2004; 101: 8751–8755.
Lee SK, Kalinowski J, Jastrzebski S, Lorenzo JA . 1,25(OH)2 vitamin D3-stimulated osteoclast formation in spleen-osteoblast cocultures is mediated in part by enhanced IL-1 alpha and receptor activator of NF-kappa B ligand production in osteoblasts. J Immunol 2002; 169: 2374–2380.
Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ . Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocrine Rev 1999; 20: 345–357.
Hobert O, Westphal H . Functions of LIM-homeobox genes. Trends Genet 2000; 16: 75–83.
Kim KK, Song SB, Kang KI, Rhee M, Kim KE . Activation of the thyroid-stimulating hormone beta-subunit gene by LIM homeodomain transcription factor Lhx2. Endocrinology 2007; 148: 3468–3476.
Hou PS, Chuang CY, Kao CF, Chou SJ, Stone L, Ho HN et al. LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res 2013; 41: 7753–7770.
Glenn DJ, Maurer RA . MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J Biol Chem 1999; 274: 36159–36167.
Perez C, Dastot-Le Moal F, Collot N, Legendre M, Abadie I, Bertrand AM et al. Screening of LHX2 in patients presenting growth retardation with posterior pituitary and ocular abnormalities. Eur J Endocrinol 2012; 167: 85–91.
Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science 2008; 319: 304–309.
Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord 2006; 7: 1–16.
Kaneko H, Arakawa T, Mano H, Kaneda T, Ogasawara A, Nakagawa M et al. Direct stimulation of osteoclastic bone resorption by bone morphogenetic protein (BMP)-2 and expression of BMP receptors in mature osteoclasts. Bone 2000; 27: 479–486.
Kim K, Kim JH, Lee J, Jin HM, Kook H, Kim KK et al. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood 2007; 109: 3253–3259.
Lee J, Kim K, Kim JH, Jin HM, Choi HK, Lee SH et al. Id helix-loop-helix proteins negatively regulate TRANCE-mediated osteoclast differentiation. Blood 2006; 107: 2686–2693.
Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med 2009; 15: 1066–1071.
Kobayashi K, Takahashi N, Jimi E, Udagawa N, Takami M, Kotake S et al. Tumor necrosis factor alpha stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J Exp Med 2000; 191: 275–286.
Queiroz-Junior CM, Bessoni RL, Costa VV, Souza DG, Teixeira MM, Silva TA . Preventive and therapeutic anti-TNF-alpha therapy with pentoxifylline decreases arthritis and the associated periodontal co-morbidity in mice. Life Sci 2013; 93: 423–428.
Chou SJ, Perez-Garcia CG, Kroll TT, O'Leary DD . Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci 2009; 12: 1381–1389.
Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 1987; 2: 595–610.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant (MRC for Gene Regulation, 2011-0030132) funded by the Korea government (MSIP) and a Grant A110703 of the Korean Health Technology R&D Project, Ministry of Health and Welfare (to JK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by JP Medema
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, J., Youn, B., Kim, K. et al. Lhx2 regulates bone remodeling in mice by modulating RANKL signaling in osteoclasts. Cell Death Differ 21, 1613–1621 (2014). https://doi.org/10.1038/cdd.2014.71
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.71
This article is cited by
-
Recent advances of NFATc1 in rheumatoid arthritis-related bone destruction: mechanisms and potential therapeutic targets
Molecular Medicine (2024)
-
LHX2 facilitates the progression of nasopharyngeal carcinoma via activation of the FGF1/FGFR axis
British Journal of Cancer (2022)
-
Selenoprotein W ensures physiological bone remodeling by preventing hyperactivity of osteoclasts
Nature Communications (2021)
-
Genomic imbalances defining novel intellectual disability associated loci
Orphanet Journal of Rare Diseases (2019)
-
mTORC1 impedes osteoclast differentiation via calcineurin and NFATc1
Communications Biology (2018)