Abstract
Progression to hormone-refractory growth of prostate cancer has been suggested to be mediated by androgen receptor (AR) gene alterations. We analyzed AR for mutations and amplifications in 21 locally recurrent prostate carcinomas treated with orchiectomy, estrogens, or a combination of orchiectomy and estramustine phosphate using fluorescence in situ hybridization, single-strand conformation polymorphism, and DNA sequence analyses. Amplification was observed in 4 of 16 (25%) and amino acid changing mutations was observed in 7 of 21 (33%) of the tumors, respectively. Two (50%) tumors with AR amplification also had missense mutation of the gene. Four of five (80%) cancers that were treated with a combination of orchiectomy and estramustine phosphate had a mutation clustered at codons 514 to 533 in the N-terminal domain of AR. In functional studies, these mutations did not render AR more sensitive to testosterone, dihydrotestosterone, androstenedione, or β-estradiol. Tumors treated by orchiectomy had mutations predominantly in the ligand-binding domain. In summary, we found molecular alterations of AR in more than half of the prostate carcinomas that recurred locally. Some tumors developed both aberrations, possibly enhancing the cancer cell to respond efficiently to low levels of androgens. Furthermore, localization of point mutations in AR seems to be influenced by the type of treatment.
Similar content being viewed by others
Introduction
Prostate cancer (CaP) is the most common male malignancy in the Western world, and its prognosis greatly depends on at which stage the disease is diagnosed. Even in the era of prostate specific antigen (PSA) screening, advanced disease is diagnosed in 20 to 40% of patients with CaP when cure by radical prostatectomy or radiotherapy is not considered possible anymore. In addition, recurrence rates of CaP after radical surgery or radiotherapy approach 25 to 50% (Dennis and Griffiths, 2000; Määttänen et al, 1999; Scardino et al, 1994). For these patients, androgen deprivation therapy (ADT) remains the only effective palliative treatment. ADT is generally achieved by either surgical or chemical castration, but for patients with widespread, metastasized disease, the cytotoxic drug estramustine phosphate (EMP) is occasionally combined with ADT (Kuhn et al, 1994; Murphy et al, 1986). Although all ADTs are initially effective, in most patients, CaP progresses within months or a few years (Dennis and Griffiths, 2000; Scardino et al, 1994). Molecular mechanisms of ADT failure are not comprehensively known. Previous studies have suggested a link between androgen receptor (AR) gene and ADT failure (Culig et al, 1993; Elo et al, 1995; Koivisto et al, 1997; Schoenberg et al, 1994; Suzuki et al, 1993, 1996; Taplin et al, 1995, 1999; Visakorpi et al, 1995; Wallen et al, 1999), and two main mechanisms by which CaP cells could adapt and sensitize AR signaling pathway for growth in low levels of androgens have been proposed. First, AR gene amplification has been shown to lead to increased expression of the AR protein and suggested to cause ADT failure (Visakorpi et al, 1995). Second, several studies suggest that AR gene mutations may influence CaP relapse (Culig et al, 1993; Elo et al, 1995; Koivisto et al, 1997; Schoenberg et al, 1994; Suzuki et al, 1993, 1996; Taplin et al, 1995, 1999; Wallen et al, 1999). Mutations can lead to changes in the receptor function by broadening the ligand specificity or increasing the transactivational activity of the receptor (Koivisto et al, 1998; Zhao et al, 2000). Additional mechanisms of AR signaling activation may involve events such as increase in coactivator levels and changes in AR phosphorylation status mediated by cytokines and peptide growth factors (Grossmann et al, 2001).
AR mutations in hormone-refractory CaP have been found in a large number of studies, but the frequency of mutations has varied from 0 to 50% (Culig et al, 1993; Elo et al, 1995; Evans et al, 1996; Gaddipati et al, 1994; Koivisto et al, 1997; Ruizeveld de Winter et al, 1994; Schoenberg et al, 1994; Suzuki et al, 1993, 1996; Taplin et al, 1995, 1999; Wallen et al, 1999). The discrepancy in mutation frequencies between the studies is considered to be due to the variability in mutation detection methodology, quality and quantity of the samples, and the heterogeneity of the clinicopathological features of the tumors. In addition, most of the reports have concentrated solely in exons 2 to 8 (Culig et al, 1993; Elo et al, 1995; Evans et al, 1996; Gaddipati et al, 1994; Koivisto et al, 1997; Ruizeveld de Winter et al, 1994; Schoenberg et al, 1994; Suzuki et al, 1993, 1996; Taplin et al, 1995, 1999), leaving exon 1 unanalyzed. In the present study, we evaluated the relevance of the AR gene in the progression of CaP during ADT achieved by orchiectomy, estrogens, or a combination of orchiectomy and EMP. We screened the AR coding region by single strand conformation polymorphism analysis and studied the AR gene copy number by fluorescence in situ hybridization (FISH) from 21 tumors. To gain additional information on the function of the mutated ARs, we performed functional studies in cell lines and modeled protein structures of two AR mutants.
Results
Twenty-one hormone-refractory transurethral resection of prostate samples taken at the time of local progression were analyzed for copy number and structural alterations of the AR gene (Table 1). Altogether, 16 AR alterations were found in 10 tumors (48%).
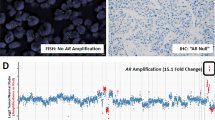
AR amplification was found in 4 of the 16 samples (25%) that were analyzed successfully by FISH. DNA sequence analysis revealed 12 changes in 9 of the 21 tumors (43%); 7 of these (58%) were located in the N-terminal domain (NTD) in exon 1 of the AR gene, and 5 (45%) were found in the ligand-binding domain (LBD). No alterations in the DNA binding domain (DBD) were found. Three tumors had a double DNA sequence change, and three tumors were found to have both AR amplification and DNA sequence change. The normal and primary tumor tissues of the mutation-positive patients were found to harbor wild-type AR, except for tumor O4, which had an R726L germ-line mutation.
Prostate Carcinomas that Recurred during a Combination of Orchiectomy and EMP
AR gene alterations were detected in four tumors (80%) from the patients who were treated with a combination of orchiectomy and EMP (Table 1). FISH analysis revealed AR amplification in tumor OE5. Four missense and one nonsense mutation were found in this therapy group. All mutations were located within the NTD between codons 514 and 533, and two of them resided at codon 524. Two mutations, G524S and W526STOP, were identified in tumor OE1. AR amplification and a mutation P533S were present in tumor OE5. Analysis of DNA from nonmalignant tissue and untreated primary tumor of the mutation-positive cases did not reveal the mutations in question.
Prostate Carcinomas that Recurred during Estrogen Treatment
Of the six carcinomas that relapsed during estrogen therapy, tumor E6 showed a high-level AR amplification (Table 1). This tumor also had two silent DNA sequence alterations: L744L and S176S. In addition, tumor E2 had a silent mutation G743G.
Prostate Carcinomas that Recurred during Orchiectomy Treatment
AR amplifications were found in two of the seven carcinomas (29%) in which FISH was successful (Table 1). In addition to AR amplification, tumor O8 had a V757I mutation, and in tumor O10, a V866M mutation was identified. Tumor O4 exhibited an R726L mutation that was also present in the patient’s germ-line DNA. This germ-line substitution has been identified before and is present in 2% of Finnish prostate cancer patients (Mononen et al, 2000). Moreover, AR CAG repeat length analysis revealed a somatic microsatellite mutation CAG24 → CAG20 in tumor O4.
Influence of the NTD Mutations on AR-Dependent Transcription
Four AR NTD missense mutations that were found in patients with CaP were recreated into mammalian expression vectors to analyze the functional consequences of the mutations. The wild-type and the P514S, G524A, G524S, and P533S mutant receptors were co-transfected into PC-3 human CaP cells (AR negative) along with a probasin promoter-driven luciferase reporter to investigate the effects of the substitutions on AR-dependent transcription. The transactivating ability of the mutants did not markedly differ from that of the wild-type AR at any of the testosterone (T) concentrations tested (0.1–100 nm). The effects of ectopic GRIP1 expression to the transcriptional activities of mutant receptors was also examined. In line with the preceding transactivation assay, all of the AR forms responded to coexpressed GRIP1; both the wild-type AR and the mutants displayed a 2- to 3-fold relative increase in the transcriptional activity in PC-3 in the presence of T or dihydro-testosterone (DHT). The activities of wild-type and mutant receptors were also studied in COS-1 cells (transformed green monkey kidney cells). With T, transcriptional activities of P514S, G534A, and G524S mutants were slightly reduced at 10 nm concentration, but their activities were comparable to that of wild-type AR at all other T concentrations. It is interesting that the mutant P533S displayed approximately 20 to 30% loss in activity compared with the wild-type AR activity at 100 nm T, and its activity was saturated already at 10 nm T. In the presence of DHT, all mutants except for P533S had activities similar to that of wild-type receptor. Again, the mutant P533S showed approximately 30% weaker transcriptional activity than the other AR forms. Similar results were obtained when a minimal promoter-driven pARE2TATA-LUC was used as a promoter (data not shown). Immunoblotting with an AR-specific antibody demonstrated that the wild-type and the mutant receptors were expressed at comparable levels in COS-1 cells.
Structural Basis of V757I and V866M Mutations
The V757 is located in the LBD in the loop between α5 and β1 helices. The substitution of the valine by isoleucine at this position results in severe clashes in all rotamer conformations with M761, and R760 from the same loop and V769 from the β2 strand. The mutation disrupts the structure of the loop and affects the conserved interaction between the M745 and G708. This association of helices α3 and α5 has been suggested to be crucial in nuclear receptor activation (Geller et al, 2000). The V866M mutation resides in the middle of α10/11 helix. The substituted methione side chain has no allowed rotamer conformations and causes unpredictable changes to the protein backbone and scaffolding.
Discussion
Molecular mechanisms of CaP relapse during ADT are largely unknown, but most of the proposed ones refer to altered AR function (Koivisto et al, 1998). In the present study, we screened all exons of the AR for mutations, as well as determined the CAG and GGC trinucleotide repeat lengths and analyzed AR gene copy number in CaPs that recurred during ADT achieved by orchiectomy, estrogens, or a combination of orchiectomy and EMP. Before this study, in only two reports the whole AR coding sequence has been analyzed in hormone-refractory CaP samples (Haapala et al, 2001; Wallen et al, 1999). Here, we detected one nonsense and seven missense mutations in one third of the tumors, and silent base changes were found in two tumors (10%). Our results are in agreement with reports by Taplin et al (1995, 1999), who discovered mutations in the AR in 31 to 50% of the bone marrow metastases that were treated by a combination of ADT and antiandrogen or with ADT alone. In the present study, somatic changes in the lengths of AR trinucleotide repeats were rare. A CAG24 → CAG20 mutation was observed in one tumor. The CAG repeat length is known to be inversely correlated with transactivation of AR, but functional and clinical results of this kind of a constriction mutation in CaP have remained unknown. In tumors that recurred during orchiectomy, all missense mutations were exclusively located in the LBD of the AR, whereas tumors that relapsed during a combination therapy of orchiectomy and EMP showed mutations in the NTD alone. None of the detected mutations was found in the corresponding untreated primary tumor, suggesting that mutations in the NTD or LBD of the AR seem to be developed and/or selected for during different types of ADT. Similar clustering of AR mutations after a particular hormonal therapy has also been observed by others (Buchanan et al, 2001; Gaddipati et al, 1994; Suzuki et al, 1996; Taplin et al, 1999). Han et al (2001) reported that 78% and 100% of the AR variants isolated from castrated and hormonally intact TRAMP mice co-localize in the NTD and LBD of the AR, respectively. Taplin et al (1999) proposed that amino acid alterations in the codon 877 are developed or selected for during a combination therapy of orchiectomy and antiandrogen. EMP, an estradiol-nitrogen mustard conjugate, which interferes with microtubules and reduces plasma levels of testosterone, is an antitumor agent used by urologists in relapsed CaP. To our knowledge, CaPs that relapsed during orchiectomy and EMP treatment have not been studied for AR mutations and copy number changes before. In the present study, five novel mutations at codons 514 to 533 in the AR NTD were detected in 80% of the tumors from the patients who were treated with a combination of orchiectomy and EMP. Our data suggest that the high incidence of mutations in the distal part of the NTD indicates that the clonal selection of tumor cells with AR NTD mutations is likely to provide CaP with a growth advantage and be associated with therapy relapse. The exact mechanisms by which mutations are driven are unknown, but a feasible hypothesis can be raised: In addition to the inhibition of the assembly of microtubules caused by the in vivo activated metabolites of EMP, EMP has been suggested to act as an androgen antagonist (Wang et al, 1998, 1999). It is interesting that Wang et al (1998) observed binding of EMP and its metabolites to the AR and showed that EMP acts, analogous to bicalutamide, antagonistically. Furthermore, they demonstrated that exposure of LNCaP cells to EMP caused transcriptional inhibition of PSA. To test whether the AR NTD missense mutations found here cause concomitant effects on the interactions between ligand-AR complexes and co-factors or possibly lead to a gain of AR activity, we evaluated their functional consequences. Our data show that the four NTD mutations did not render AR more sensitive to T and DHT in human CaP cells. Moreover, the responses of the mutants to β-estradiol and adrenal androgen androstenedione did not markedly differ from that of the wild-type receptor (data not shown). However, the mutant P533S displayed a slight reduction in its transactivation activity with T in COS-1 cells. AR NTD mutations in the orchiectomy and EMP therapy group clustered within amino acids 514 to 533, which may reside in binding surface for the coactivators, such as GRIP1/TIF2 (Irvine et al, 2000) and Cyclin E (Yamamoto et al, 2000). Several coactivators, such as ARA160, BRCA1, TFIIH, and TRAM-1 are known to interact with NTD, but specific binding sequence of the domain has not been characterized.
A large percentage of recurrent CaPs express high levels of GRIP1/TIF2 and/or SRC1 (Gregory et al, 2001). However, the AR NTD mutants found in this study did not significantly differ from the wild-type AR in their responses to ectopically expressed GRIP1. PIAS1 represents another type of AR coactivator that has been shown to interact, in part, with the NTD (Tan et al, 2000). In addition, PIAS1 modulated the activities of the mutants in a manner that was comparable to that of wild-type receptor (data not shown). Our current data showing that the activities of the NTD mutants do not markedly differ from that of the wild-type AR under transient expression conditions does not rule out the possibility that these mutations alter AR function in vivo. Moreover, it is possible that the most important coactivator proteins for AR function in prostate remain to be identified. Forty different mutations identified in the AR LBD in clinical CaP have been listed in the Androgen Receptor Mutation Database (www.mcgill.ca/androgendb). In the present study, two somatic missense mutations, V757I and V866M, were found in the LBD. The V757I variant has not been described before, but a valine to alanine substitution in the codon 757 has been reported in a metastatic CaP (Marcelli et al, 2000). Amino acid 757 is localized in a highly conserved region of the LBD, and in accordance with our structure modeling, V757I mutation is likely to distort the AR protein backbone and at least locally the fold of the protein. Therefore, it is predicted that the ligand-binding capacity of the V757I variant is altered when compared with wild-type receptor, but functional studies are required to demonstrate it.
The V866M variant has been previously found in CaP and androgen insensitivity syndrome (Androgen Receptor Mutation Database). Four studies have measured androgen binding to the AR V866M mutant and found normal binding capacity with reduced ligand binding affinity in patients with androgen insensitivity syndrome (Hiort et al, 1998; Kazemi-Esfarjani et al, 1993; Lubahn et al, 1989; Weidemann et al, 1996), whereas one report (Ahmed et al, 2000) found no binding capacity. DHT has remained the only ligand tested in these studies, and whether the V866M substitution leads to alteration of steroid specificity is unclear. Structural analysis of the V866M mutant showed that the valine to methionine substitution causes a conformational change, which is likely to disrupt the ligand-binding pocket of AR.
In addition to structural changes of AR, the AR gene amplification is another suggested mechanism of ADT relapse in CaP (Visakorpi et al, 1995). We previously reported AR amplification and overexpression in 30% of the CaPs that recurred during ADT (Bubendorf et al, 1999; Koivisto et al, 1997; Visakorpi et al, 1995). In the present study, the prevalence of AR gene amplification was 25%, which is in line with reports from other laboratories (Kaltz-Wittmer et al, 2000; Miyoshi et al, 2000). The tumors with amplification were equally distributed among the therapy groups, suggesting that orchiectomy, estrogen therapy, and a combination therapy of orchiectomy and EMP are able to cause such a reduction in intraprostatic DHT levels that the growth of malignant cells is disturbed and only cell clones with selective growth advantages, such as extra AR copies, are survived under the androgen-deficient milieu. In addition, missense mutations were detected in 50% of the tumors with AR amplification. The coexistence of AR gene mutations and amplification in the same tumor has been previously reported (Koivisto et al, 1997; Wallen et al, 1999). Moreover, there are two previous reports on the tumors with multiple AR mutations in hormone-refractory prostate cancers (Gaddipati et al, 1994; Taplin et al, 1995). Thus, the coexistence of AR mutation and amplification is likely to provide a synergistic growth advantage for prostate cancer cells in hormonally altered cellular environment.
In summary, we found molecular aberrations of the AR gene in half of the CaPs that recurred locally. Moreover, our results suggest that the selective pressure caused by various types of ADT may determine the nature of mutations in the AR. The collocation of AR mutations at the codons 514 to 533 in CaP from patients who received cytotoxic agent EMP represents a possible molecular mechanism for the development of therapy-resistant CaP. Identification and further characterization of these mechanisms may have an impact on the development of new regimens for the medical treatment of hormone-refractory CaP.
Materials and Methods
Patients and Tumor Specimens
Twenty-one patients with advanced CaP were treated at the Tampere University Hospital in Finland. All patients experienced a local tumor recurrence, as evidenced by symptoms of urethral obstruction and an increase of serum prostatic acid phosphatase (before 1991) or serum PSA levels (after 1991). TNM stage of the tumors at the time of diagnosis and histological differentiation of the recurrent tumors are summarized in Table 1. The clinical data were collected from the patient records. Formalin-fixed paraffin-embedded transurethral resection of prostate samples were available from all recurrent tumors. For DNA extraction and tissue array, representative tumor areas that contained >80% of malignant cells were selected by histopathological examination of hematoxylin and eosin–stained slides. Nonmalignant tissue was obtained from all mutation-positive patients, and untreated primary tumor sample was available from 2 of 8 cases (25%) that contained somatic AR mutation (Table 1). The research protocol was approved by the Ethical Committee of the Tampere University Hospital.
Analysis of Gene Amplification by FISH
FISH was performed on sections of the formalin-fixed tumor samples on a tissue array (Andersen et al, 2001; Kononen et al, 1998). Four samples were previously analyzed for AR gene copy number (Bubendorf et al, 1999) and excluded from the present tissue array block. Five-micrometer-thick sections were transferred onto TechMate slides (Dako A/S, Glostrup, Denmark) and fixed overnight at 56° C. Two-color FISH was performed using Spectrum Orange-labeled AR probe (Vysis, Downer’s Grove, Illinois) and FITC-labeled probe (DXZ1/BamX7) specific for the centromere of the X chromosome. Hybridization mixture contained 1 μl of both probes and 8 μl of hybridization buffer (Vysis). After overnight hybridization, the slides were washed and counterstained with antifade solution (Vectashield; Vector Laboratories, Burlingame, California) containing 0.2 μm of 4′,6-diamidino-2-phenylindole. The signals were scored with a Leica DMRB (Leica Microsystems, Wetzlar, Germany) fluorescence microscope equipped with a double-band pass filter using ×100 objective. At least 50 cells were scored per sample. The number of AR signals was evaluated in relation to the DXZ1 signals, and the AR gene was regarded as amplified when the copy number of the AR gene was at least 3-fold higher than that of the DXZ1.
PCR and Single Strand Conformation Polymorphism Analysis
DNA was isolated from paraffin-embedded tumor specimens using standard phenol-chloroform extraction. PCR was performed in a volume of 10 μl using GeneAmp PCR System 9700 (Applied Biosystems Inc., Foster City, California). Sample DNA (25 ng) was amplified using 35 cycles of PCR (94° C for 30 seconds, 55–60° C for 30 seconds, and 72° C for 45 seconds). Reaction mixture contained 1× PCR buffer (1.5 mm of MgCl2), 20 μm of each dNTPs, 0.6 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), 0.4 μm of each primer, and 0.4 μCi of [α33P]-dCTP (>2,500 Ci/mmol). PCR fragments were subsequently heat-denatured and run for 15 to 24 hours at 800 V in 0.5× and 0.8× mutation detection enhancement (MDE) gels (BMA, Rockland, Maine) containing 1% glycerol. After electrophoresis, the gels were dried and exposed to Kodak BioMax MR film for 1 day. The entire AR gene from normal human DNA and exon 8 from LNCaP cell line, containing a T877A mutation, were amplified and run as controls along with the samples. When band shift was noticed, the single strand conformational polymorphism procedure was repeated and the aberrant band was cut from the gel, reamplified in 50 μl, and sequenced.
Primers
Primers reported previously for exon 1 of the AR gene by Tilley et al (1996) were used with the exception of region 40 to 249 (coordinates from Lubahn et al, 1989) that was amplified with 5′-GGGTAAGGGAAGTAGGTGGA-3′ and 5′-CAGCAGCAGCAAACTGGC-3′ and the region 1114 to 1309 was amplified with 5′-TACAAGTCCGGAGCACTGGA-3′ and 5′-ATGCAGGCTCGCCAGGTC-3′. Primers for exons 2 to 8 were published previously (Koivisto et al, 1997), except that exons 4, 7, and 8 were amplified with the following pairs of primers: 5′-GCATTGTGTGT-TTTTGACCACTGATG-3′ and 5′-TGCAAAGGAGTTGGGCTGGTTG-3′; 5′-TCTAATGCTCCTTCGTGGGCA-3′ and 5′-CTCTATCAGGCTGTTCTCCCT-3′; 5′-TCAACCCTGTTTTTCTCCCT-3′ and 5′-TTTCCCAGAAAGGATCTTGG-3′, respectively.
Repeat Length Analysis
PCR amplification of the CAG and GGC repeats was carried out using conditions described previously (Mitchell et al, 2000) using FastStart DNA polymerase kit (Roche Molecular Biochemicals, Mannheim, Germany). Primers were 5′-GCCTGTTGAACTCTTCTGAGC-3′ and 5′-GCTGTGAAGGTTGCTGTTCCTC-3′ for CAG and 5′-ACAGCCGAAGAAGGCCAGTTGT-3′ and 5′-CAGGTGCGGTGAAGTCGCTTTCCT-3′ for GGC. Labeled PCR products were generated by adding [α33P]-dCTP and denatured by heating in a formamide dye and then analyzed by electrophoresis in a 6% polyacrylamide/8M urea at 70 W power. For each run, the five samples of previously determined sizes were included as controls.
DNA Sequencing
The sequencing was carried out with the fmol DNA Cycle Sequencing System (Promega, Madison, Wisconsin) using the same primers as for PCR. All potential mutations were sequenced on both DNA strands. Sequencing gel electrophoresis was performed using standard conditions.
Plasmid Constructions
The individual NTD point mutations were incorporated into AR using overlap PCR (sequences of the primers available upon request). PCR was performed for 14 cycles using Accutaq LA DNA polymerase (Sigma) in accordance with the manufacturer’s instructions. The resulting fragments were digested with KpnI and Tth111I and transferred into a corresponding restriction site of pSG5-hAR (Adeyemo et al, 1993). Mutation incorporation was checked by sequencing both strands using the Pharmacia ALFexpress DNA sequencing system (Amersham Pharmacia Biotech, Buckinghamshire, UK). pPB(−285/+32)-LUC containing nucleotides −285 to 32 of the rat probasin promoter driving firefly luciferase (LUC) coding region and pARE2TATA-LUC that contains two AREs (from the first intron of the rat C3 gene) in front of minimal TATA sequence have been described (Palvimo et al, 1996).
Transfections and Reporter Gene Assays
PC-3 cells (from American Type Culture Collection, Manassas, Virginia) were maintained in Nutrient Mixture F-12 (HAM) containing penicillin (25 U/ml), streptomycin (25 U/ml), and 7% fetal bovine serum and supplemented with l-glutamine (250 mg/L). Transfections were performed using FuGENE 6 reagent (Roche) according to the manufacturer’s instructions. In brief, 60 × 103 PC-3 cells were seeded on 12-well plates 24 hours before transfection. Four hours before transfection, cells received fresh medium containing 7% charcoal-stripped fetal bovine serum and l-glutamine (250 mg/L). A total of 200 ng of reporter plasmid, 50 ng of internal control pCMVβ (Clontech), and 20 ng of pSG5-AR expression constructs were transfected. Eighteen hours after transfection, the cells received fresh medium containing 7% charcoal-stripped fetal bovine serum and indicated concentrations of T, DHT, or vehicle. After a 30-hour culture, the cells were harvested and lysed in reporter lysis buffer (Promega) and the LUC and β-galactosidase activities were assayed (Palvimo et al, 1996). Experiments using COS-1 cells were performed essentially as described (Thompson et al, 2001) using 200 ng of reporter plasmid, 50 ng of pCMVβ, and 2 ng of pSG5-AR expression plasmid.
Immunoblot Analysis of Extracts from Transfected Cells
Transfected COS-1 cells were washed twice with phosphate-buffered saline, and the pellets were resuspended in SDS-PAGE sample buffer, heated at 95° C for 5 minutes, and analyzed on a 10% SDS-polyacrylamide gel. The proteins were transferred onto a nitrocellulose membrane (Amersham Pharmacia Biotech). Membranes were then immunoblotted with an antiserum K333 raised against full-length rat AR (Karvonen et al, 1997). The membranes were subsequently incubated with horseradish phosphatase-conjugated goat anti-rabbit immunoglobulin G antibody (Zymed Laboratories, South San Francisco, California), and immunocomplexes were detected using the ECL Western blot reagents (Amersham Pharmacia Biotech) according to the manufacturer’s instructions.
Structural Analyses of the V757I and V866M Mutations
The V757I and V866M mutations were analyzed on the basis of the crystal structure of the ligand-binding domain of human androgen receptor (Matias et al, 2000; PDB code 1E3G). The rotatable side chains were created with PREKIN 5.93, and an automated sampling of torsional angles was done with Autobondrot procedure under PROBE 2.80 as previously described (Word et al, 2000). The χ angles were increased by 5° in each step. The acceptable conformations for a mutated side chain have a total score above −1.0 (Lovell et al, 2000). The contact surfaces showing the steric clashes as well as the positive van der Waals interactions of a mutated side chain with the surrounding residues were analyzed by using Mage 5.93. All of the programs used were obtained from kinemage.biochem.duke.edu.
References
Adeyemo O, Kallio PJ, Palvimo JJ, Kontula K, and Jänne OA (1993). A single-base substitution in exon 6 of the androgen receptor gene causing complete androgen insensitivity: The mutated receptor fails to transactivate but binds to DNA in vitro. Hum Mol Genet 2: 1809–1812.
Ahmed SF, Cheng A, Dovey L, Hawkins JR, Martin H, Rowland J, Shimura N, Tait AD, and Hughes IA (2000). Phenotypic features androgen receptor binding and mutational analysis in 278 clinical cases reported as androgen insensitivity syndrome. J Clin Endocrinol Metab 85: 658–665.
Andersen CL, Hostetter G, Grigoryan A, Sauter G, and Kallioniemi A (2001). Improved procedure for fluorescence in situ hybridization on tissue microarrays. Cytometry 45: 83–86.
Bubendorf L, Kononen J, Koivisto P, Schraml P, Moch H, Gasser TC, Willi N, Mihatsch MJ, Sauter G, and Kallioniemi OP (1999). Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res 59: 803–806.
Buchanan G, Greenberg NM, Scher HI, Harris JM, Marshall VR, and Tilley WD (2001). Collocation of androgen receptor gene mutations in prostate cancer. Clin Cancer Res 7: 1273–1281.
Culig Z, Hobisch A, Cronauer MV, Cato AC, Hittmair A, Radmayr C, Eberle J, Bartsch G, and Klocker H (1993). Mutant androgen receptor detected in an advanced-stage prostatic carcinoma is activated by adrenal androgens an progesterone. Mol Endocrinol 7: 1541–1550.
Dennis LJ and Griffiths K (2000). Endocrine treatment in prostate cancer. Semin Surg Oncol 18: 52–74.
Elo JP, Kvist L, Leinonen K, Isomaa V, Henttu P, Lukkarinen O, and Vihko P (1995). Mutated human androgen receptor gene detected in a prostatic cancer patients is also activated by estradiol. J Clin Endocrinol Metab 80: 3494–3500.
Evans BAJ, Harper ME, Daniels CE, Watts CE, Matenhelia S, Green J, and Griffiths K (1996). Low incidence of androgen receptor gene mutations in human prostatic tumors using single strand conformation polymorphism analysis. Prostate 28: 162–171.
Gaddipati JP, McLeod DG, Heidenberg HB, Sesterhenn IA, Finger MJ, Moul JW, and Srivastava S (1994). Frequent detection of codon 877 mutation in the androgen receptor gene in advanced prostate cancers. Cancer Res 54: 2861–2864.
Geller DS, Farhi A, Pinkerton N, Fradley M, Moritz M, Spitzer A, Meinke G, Tsai FT, Sigler PB, and Lifton RP (2000). Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science 289: 23–26.
Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, and Wilson EM (2001). A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res 61: 4315–4319.
Grossmann ME, Huang H, and Tindall DJ (2001). Androgen receptor signaling in androgen-refractory prostate cancer. J Natl Cancer Inst 93: 1687–1697.
Haapala K, Hyytinen E-R, Roiha M, Laurila M, Rantala I, Helin HJ, and Koivisto PA (2001). Androgen receptor alterations in prostate cancer relapsed during a combined androgen blockade by orchiectomy and bicalutamide. Lab Invest 81: 1647–1651.
Han G, Foster BA, Mistry S, Buchanan G, Harris JM, Tilley WD, and Greenberg NM (2001). Hormone status selects for spontaneous somatic androgen receptor variants that demonstrate specific ligand and cofactor dependent activities in autochthonous prostate cancer. J Biol Chem 276: 11204–11213.
Hiort O, Sinnecker GH, Holterhus PM, Nitsche EM, and Kruse K (1998). Inherited and de novo androgen receptor gene mutations: Investigation of single-case families. J Pediatr 132: 939–943.
Irvine RA, Ma H, Yu MC, Ross RK, Stallcup MR, and Coetzee GA (2000). Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum Mol Genet 9: 267–274.
Kaltz-Wittmer C, Klenk U, Glaessgen A, Aust DE, Diebold J, Lohrs U, and Baretton GB (2000). FISH analysis of gene aberrations (MYC, CCND1, ERBB2, RB and AR) in advanced prostatic carcinomas before and after androgen deprivation therapy. Lab Invest 80: 1455–1464.
Karvonen U, Kallio PJ, Jänne OA, and Palvimo JJ (1997). Interaction of androgen receptors with androgen response element in intact cells. Roles of amino- and carboxyl-terminal regions and the ligand. J Biol Chem 272: 15973–15979.
Kazemi-Esfarjani P, Beitel LK, Trifiro M, Kaufman M, Rennie P, Sheppard P, Matusik R, and Pinsky L (1993). Substitution of valine-865 by methionine or leucine in the human androgen receptor causes complete or partial androgen insensitivity respectively with distinct androgen receptor phenotypes. Mol Endocrinol 7: 37–46.
Koivisto P, Kolmer M, Visakorpi T, and Kallioniemi OP (1998). Androgen receptor gene and hormonal therapy failure of prostate cancer. Am J Pathol 152: 1–9.
Koivisto P, Kononen J, Palmberg C, Hyytinen E, Tammela T, Trapman J, Isola J, Visakorpi T, and Kallioniemi O-P (1997). Androgen receptor gene amplification: A possible molecular mechanism for failure of androgen deprivation therapy in prostate cancer. Cancer Res 57: 314–319.
Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, and Kallioniemi O-P (1998). Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4: 844–847.
Kuhn MW, Weissbach L, and Hinke A (1994). Primary therapy of metastatic prostate carcinoma with depot gonadotropin-releasing hormone analogue goserelin versus estramustine phosphate. The Prostate Cancer Study Group. Urology 43 (Suppl): 61–67.
Lovell SC, Word JM, Richardson JS, and Richardson DC (2000). The penultimate rotamer library. Proteins 40: 389–408.
Lubahn DB, Brown TR, Simental JA, Higgs HN, Migeon CJ, Wilson EM, and French FS (1989). Sequence of the intron/exon junctions of the coding region of the human androgen receptor gene and identification of a point mutation in a family with complete androgen insensitivity. Proc Natl Acad Sci USA 86: 9534–9538.
Määttänen L, Auvinen A, Stenman UH, Rannikko S, Tammela T, Aro J, Juusela H, and Hakama M (1999). European randomized study of prostate cancer screening: First-year results of the Finnish trial. Br J Cancer 79: 1210–1214.
Marcelli M, Ittmann M, Mariani S, Sutherland R, Nigam R, Murthy L, Zhao Y, DiConcini D, Puxeddu E, Esen A, Eastham J, Weigel NL, and Lamb DJ (2000). Androgen receptor mutations in prostate cancer. Cancer Res 60: 944–949.
Matias PM, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, Otto N, Joschko S, Scholz P, Wegg A, Basler S, Schafer M, Egner U, and Carrondo MA (2000). Structural evidence for ligand specificity in the binding domain of the human androgen receptor: Implications for pathogenic gene mutations. J Biol Chem 275: 26164–26171.
Mitchell S, Abel P, Ware M, Stamp G, and Lalani E (2000). Phenotypic and genotypic characterization of commonly used human prostatic cell lines. BJU Int 85: 932–944.
Miyoshi Y, Uemura H, Fujinami K, Mikata K, Harada M, Kitamura H, Koizumi Y, and Kubota Y (2000). Fluorescence in situ hybridization evaluation of c-myc and androgen receptor gene amplification and chromosomal anomalies in prostate cancer in Japanese patients. Prostate 43: 225–232.
Mononen N, Syrjäkoski K, Matikainen M, Tammela TL, Schleutker J, Kallioniemi OP, Trapman J, and Koivisto PA (2000). Two percent of Finnish prostate cancer patients have a germ-line mutation in the hormone-binding domain of the androgen receptor gene. Cancer Res 60: 6479–6481.
Murphy GP, Huben RP, and Priore R (1986). Results of another trial of chemotherapy with and without hormones in patients with newly diagnosed metastatic prostate cancer. Urology 28: 36–40.
Palvimo JJ, Reinikainen P, Ikonen T, Kallio PJ, Moilanen A, and Jänne OA (1996). Mutual transcriptional interference between RelA and androgen receptor. J Biol Chem 271: 24151–24156.
Ruizeveld de Winter JA, Janssen PJ, Sleddens HM, Verleun-Mooijman MC, Trapman J, Brinkmann AO, Santerse AB, Schroder FH, and van der Kwast TH (1994). Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am J Pathol 144: 735–746.
Scardino PT, Beck JR, and Miles BJ (1994). Conservative management of prostate cancer. N Engl J Med 330: 1831.
Schoenberg MP, Hakimi JM, Wang S, Bova GS, Epstein JI, Fiscbeck KH, Isaacs WB, Walsh PC, and Barrack ER (1994). Microsatellite mutation (CAG24♦8) in the androgen receptor gene in human prostate cancer. Biochem Biophys Res Comm 198: 74–80.
Suzuki H, Akakura K, Komiya A, Aida S, Akimoto S, and Shimazaki J (1996). Codon 877 mutation in the androgen receptor gene in advanced prostate cancer: Relation to antiandrogen withdrawal syndrome. Prostate 29: 153–158.
Suzuki H, Sato N, Watabe Y, Masai M, Seino S, and Shimazaki J (1993). Androgen receptor gene mutations in human prostate cancer. J Steroid Biochem Mol Biol 46: 759–765.
Tan I, Hall SH, Hamil KG, Grossman G, Petrusz P, Liao L, Shuai K, and French ES (2000). Protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) is a nuclear receptor coregulator expressed in human testis. Mol Endocrinol 14: 14–26.
Taplin ME, Bubley GJ, Ko YJ, Small EJ, Upton M, Rajeshkumar B, and Balk SP (1999). Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res 59: 2511–2515.
Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, and Keer HN (1995). Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med 332: 1393–1398.
Thompson J, Saatcioglu F, Jänne OA, and Palvimo JJ (2001). Disrupted amino- and carboxyl-terminal interactions of the androgen receptor are linked to androgen insensitivity. Mol Endocrinol 15: 923–935.
Tilley WD, Buchanan G, Hickey TE, and Bentel JM (1996). Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin Cancer Res 2: 277–285.
Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C, Palotie A, Tammela T, Isola J, and Kallioniemi O-P (1995). In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 9: 401–406.
Wallen MJ, Linja M, Kaartinen K, Schleutker J, and Visakorpi T (1999). Androgen receptor gene mutations in hormone-refractory prostate cancer. J Pathol 189: 559–563.
Wang LG, Liu XM, Kreis W, and Budman DR (1998). Androgen antagonistic effect of estramustine phosphate (EMP) metabolites on wild-type and mutated androgen receptor. Biochem Pharmacol 55: 1427–1433.
Wang LG, Liu XM, Kreis W, and Budman DR (1999). Phosphorylation/dephosphorylation of androgen receptor as a determinant of androgen agonistic or antagonistic activity. Biochem Biophys Res Commun 259: 21–28.
Weidemann W, Linck B, Haupt H, Mentrup B, Romalo G, Stockklauser K, Brinkmann AO, Schweikert HU, and Spindler KD (1996). Clinical and biochemical investigations and molecular analysis of subjects with mutations in the androgen receptor gene. Clin Endocrinol 45: 733–739.
Word JM, Bateman RC Jr, Presley BK, Lovell SC, and Richardson DC (2000). Exploring steric constraints on protein mutations using MAGE/PROBE. Protein Sci 11: 2251–2259.
Yamamoto A, Hashimoto T, Kohri K, Ogata E, Kato S, Ikeda K, and Nakanishi M (2000). Cyclin E as a coactivator of the androgen receptor. J Cell Biol 150: 873–880.
Zhao XY, Malloy PJ, Krishnan AV, Swami S, Navone NM, Peehl DM, and Feldman D (2000). Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med 6: 703–706.
Acknowledgements
This study was financially supported by the Medical Research Fund of Tampere University Hospital, the Lahtikari Foundation, the Finnish Cancer Institute (to PAK and E-RH), and the Pirkanmaa Regional Fund of the Finnish Cultural Foundation (to KH) and Finnish Academy (to MV, JJP, and OAJ).
We thank Mrs. Sinikka Kekkonen and Mrs. Lila Nikkola for skillful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
KH and JT contributed equally to this work.
Rights and permissions
About this article
Cite this article
Hyytinen, ER., Haapala, K., Thompson, J. et al. Pattern of Somatic Androgen Receptor Gene Mutations in Patients with Hormone-Refractory Prostate Cancer. Lab Invest 82, 1591–1598 (2002). https://doi.org/10.1097/01.LAB.0000038924.67707.75
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.LAB.0000038924.67707.75
This article is cited by
-
Androgen receptor signaling in prostate cancer
Cancer and Metastasis Reviews (2014)
-
AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression
Oncogene (2012)
-
Androgen receptor signaling and mutations in prostate cancer
Asian Journal of Andrology (2010)
-
Targeted therapy of cancer: new roles for pathologists—prostate cancer
Modern Pathology (2008)
-
Positron tomographic assessment of androgen receptors in prostatic carcinoma
European Journal of Nuclear Medicine and Molecular Imaging (2005)