Abstract
Our laboratory has recently demonstrated constitutive activation of the Notch signaling pathway in Kaposi's sarcoma tumor cells. As endothelial cells (EC) are believed to be the progenitor of these tumor cells, this study was designed to examine the effect of Notch activation on normal human EC. Recent reports suggest Notch activation induces EC growth arrest, and that this growth arrest may be linked to the establishment or maintenance of EC quiescence, the phenotype seen in contact-inhibited EC lining the vasculature. To gain further insight into Notch activation and quiescence, we first confirmed that Notch activation induced EC growth arrest. Next, we examined Notch activation in confluent, growth arrested EC (mimicking the cells lining the vasculature). In contrast to previous reports, we found confluent EC possess lower levels of activated Notch compared to proliferating control cells. Interestingly, these cells express elevated levels of Hes-1 protein (an immediate downstream target of Notch signaling) despite decreased Notch activation. Under these conditions, Hes-1 expression was mediated, at least in part, by a Notch-independent mechanism involving c-jun N-terminal protein kinase (JNK) signaling. This is the first report, to our knowledge, that JNK signaling can modulate Hes-1 expression in a Notch-independent manner.
Similar content being viewed by others
Main
The Notch pathway is an evolutionarily conserved intercellular signaling mechanism that plays a prominent role in cell fate decisions including proliferation, differentiation, and survival.1 Notch receptors (Notch-1, -2, -3, -4) interact with ligands (Jagged-1, -2, and Delta-like (Dll)-1, -3, -4) on adjacent cells triggering proteolytic cleavage of the receptor by the presenilin/gamma-secretase complex. This releases the intracellular domain of the Notch receptor (NIC), which translocates to the nucleus and binds CBF-1 (also termed CSL or RBP-Jκ), a DNA binding protein.2 In the absence of NIC CBF-1 represses gene transcription by binding with the histone–deacetylase complex (SMRT/sin3/HDAC-1); however, binding of NIC to CBF-1 displaces the repressor complex and recruits nuclear coactivators converting CBF-1 to a transcriptional activator. Notch-mediated transcription results in the expression of various target genes, including the Hes (Hairy/Enhancer of Split) and Hey (Hairy/Enhancer of Split related with YRPW, also known as HesR, HRT, and HERP) families of transcription factors.3
Hes (-1, -3, -5) and Hey (-1, -2, -3) family members have been identified as immediate downstream targets of Notch activation. These proteins are transcriptional repressors that act by negatively regulating expression of target genes such as tissue-specific transcriptional activators. Hes and Hey proteins are structurally similar as they each contain a basic helix–loop–helix (bHLH) domain, an orange domain, and a tetrapeptide motif; however, they have distinct mechanisms of action. Hes interacts with TLE/Groucho through the tetrapeptide motif while Hey engages the mSin3 complex through the bHLH domain.3, 4 These proteins can form both homo- and heterodimers, which is believed to extend their individual repression activity.3
Previous studies indicate that Notch signaling is critical in the formation and maintenance of the vasculature. Endothelial cells (EC) express Notch-1, -2, and -4 as well as the ligands Jagged-1, -2, and Dll-1, -4.5, 6, 7, 8, 9, 10 Furthermore, targeted deletion of notch-1, notch-4, dll-1, jagged-1, or jagged-2 in mice results in vascular defects ranging from minor disruption of capillary branching to embryonic lethality characterized by severe hemorrhage.11, 12 Interestingly, constitutive Notch-4 activation also results in vascular defects, indicating the importance of tightly controlled Notch signaling during vascular development.13
We have recently demonstrated constitutive Notch activation in Kaposi's sarcoma (KS), the most common tumor in AIDS patients.14, 15 These studies showed that blockade of Notch signaling resulted in growth inhibition and apoptosis of KS tumor cells. As EC are the likely progenitor of KS tumor cells, we were intrigued by reports indicating Notch activation induces growth arrest in EC, and that Notch activation may be linked to the establishment or maintenance of EC quiescence as seen in the contact-inhibited EC lining normal vasculature.16, 17, 18 Therefore, this study was designed to gain insight into a possible role for Notch activation in EC quiescence. In contrast to earlier reports, we found elevated levels of Hes-1 in confluent, growth arrested EC (mimicking the vasculature) were not associated with Notch activation, but instead appeared to be mediated, at least in part, by a Notch-independent mechanism involving c-jun N-terminal protein kinase (JNK) signaling.
Materials and methods
Cell Culture
Human umbilical vein EC were isolated from freshly obtained human umbilical cords by treatment with collagenase.19 EC were plated on gelatin coated tissue culture dishes and maintained in EGM-2MV (Cambrex, Walkersville, MD, USA) using standard tissue culture techniques. The Phoenix-Ampo retroviral packaging cells were obtained from the American Type Culture Collection with permission from Garry P Nolan.20 The packaging cells were cultured in Dulbecco modified Eagles medium containing 10% fetal bovine serum (FBS). In a majority of the experiments, cells were actively proliferating (50–60% confluent) when samples were harvested and analyzed. For studies on confluent EC, samples were harvested 96 h after visual confluence was reached. To ensure nutrient availability was equivalent, media changes were performed simultaneously on both proliferating and confluent cultures. The following compounds from EMD Biosciences (San Diego, CA, USA) were used in signaling experiments: FTI-277 (Ras inhibitor), LY294002 (phosphoinositide kinase-3 (PI3K) inhibitor), PD98059 (mitogen-activated protein kinase (MAPK) inhibitor), SB203580 (p38 inhibitor), GF109203X (protein kinase C (PKC) inhibitor), NF-κB activation inhibitor, extracellular signal-regulated kinase (ERK) inhibitor peptide II, JNK inhibitor I, and JNK inhibitor II (SP600125).
Retroviral Expression Vectors and Transductions
The LZRS retroviral expression vector has been previously used in our laboratory.21, 22 Sequences encoding for NIC-1, NIC-2, NIC-4, and Jagged-1 were subcloned into LZRS and expression verified by Western blot. The Phoenix packaging cell line was transfected with the constructs using standard CaCl2 and 2 × Hanks balanced salt solution methodologies. After overnight incubation, fresh media was added, and the cells incubated at 32°C for an additional 24–48 h. The retrovirus-containing supernatants were collected, filtered to remove contaminating cells and stored at −80°C. EC (2 × 105 cells/well in six-well plates) were transduced with 1 ml of viral supernatant in the presence of 8 μg polybrene for 1 h with centrifugation. The cells were incubated for 48 h before testing for transduction efficiency and use in experiments. Using this protocol, we can reproducibly transduce >85% of ECs (range: 87.4–93%).21 Therefore, transduced cells were used as heterogenous bulk cultures and were not selected with antibiotics.
Western Blot Analysis
Whole cell extracts were prepared by lysing cells in CHAPS buffer containing a mixture of protease inhibitors as described.22 Protein concentration was determined using a Bradford Assay (Bio-Rad Laboratories, Hercules, CA, USA). Protein (50 μg) was loaded on sodium dodecyl sulfate-polyacrylamide gels, transferred to an Immobilon-P membrane and blocked with 5% powdered milk in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, 0.01% Tween-20). The membrane was then incubated with primary antibodies diluted in 2.5% powdered milk in TBST, washed extensively and incubated with HRP-conjugated species specific secondary antibodies (Amersham Biosciences, Piscataway, NJ, USA). Proteins were visualized with ECL reagents (Amersham Biosciences) according to the manufacturer's instructions. Even loading of proteins was confirmed by Ponceau S staining and detection of the housekeeping protein, actin, on each blot. The antibodies against Notch-1 (bTan20), Notch-2 (C651.6DbHN), and Jagged-1 (TS1.15H), developed by Dr S Artavanis-Tsakonas were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa (Iowa City, IA, USA). Antibodies directed against Notch-4, Delta1, Delta4 were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA, USA) and those detecting JNK, p38, ERK1/2, MEK1/2 and their respective phosphorylated forms were obtained from Cell Signaling Technologies. Antibodies directed against actin (MP Biomedicals, Irvine, CA, USA), Hes-1 (BD Biosciences, San Jose, CA, USA), and Hey-1 (Chemicon International, Temecula, CA, USA) were also used in this study. Differences in protein expression were determined by densitometry analysis using Scion Image Software (Scion Corporation, Frederick, MD, USA). Western blot experiments were repeated at least twice to confirm the results.
Transfections and Luciferase Assays
The Hes-1A/B luciferase reporter construct, containing the −194 to +160 promoter fragment of the Hes-1 gene inserted upstream of the luciferase gene in pGL2, was the gift of Dr S Sisodia (University of Chicago, Chicago, IL, USA).23 The Hey-1 construct contains approximately 3 kb of the presumed promoter region (−2839 to +87) of Hey-1 upstream of the luciferase gene in pLuc and was the gift of Dr M Gessler (University of Wuerzburg, Wuerzburg, Germany). Cells (2 × 104 cells/well in a 24-well plate) were transfected using Cytopure transfection reagent (MP Biomedicals) following the manufacturer's instructions. At 24 h after transfection, luciferase activity was determined using Bright-N-Glo luciferase reagent.24 The results were corrected for transfection efficiency, and individual experiments were normalized to each other using the baseline luciferase activity in cells transfected with the empty luciferase vector (pGL2 or pLuc) as previously described.24 Experiments were performed in triplicate.
RNA Interference
A validated siRNA duplex to block JNK expression and a negative control siRNA were purchased from Qiagen (Valencia, CA, USA). RNA duplexes were suspended at 20 μM in suspension buffer. The siRNA was mixed with 40 μl HiPerfect Transfection Reagent (Qiagen) and diluted in complete media for a final concentration of 5 nM. EC were treated with the transfection mixture for 48 h after which total protein was harvested for Western blot analysis.
Cell Cycle Analysis
DNA/propidium iodide (DNA/PI) staining was performed using standard methodologies. 1 × 106 cells were permeabilized with 100% ethanol in the presence of 15% FBS. The cells were washed and then treated for 15 min at 37°C with 10 μg/ml RNAse. 5 μg/ml PI was added, and the cells incubated for 1 h at 4°C before analysis by flow cytometry using a Coulter Epics MCL flow cytometer with 10 000 cells analyzed per gated determination.
Proliferation Assays
Cell proliferation was quantitated using a WST-1 assay, a highly sensitive, colorimetric alternative to 3H-thymidine incorporation assays. Briefly, EC (4 × 103 cells/well in a 96-well plate) were treated as indicated in the text. After 24 h, 10 μl of WST-1 reagent (Roche Applied Science, Indianapolis, IN, USA) was added per well. After a 4 h incubation, the plate was shaken for 1 min, and color development measured on a microplate reader at 450 nm.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
Complementary DNA was synthesized by reverse transcription of total RNA using oligodT as a primer. Quantitative Polymerase Chain Reaction (PCR) was then performed using a GeneAmp 5700 sequence detection system (Applied Biosystems, Foster City, CA, USA) with Quantitect SYBR Green PCR reagents and Hes-1 Quantitect Primers from Qiagen following the manufacturer's instructions. Each sample was also run under identical conditions with GAPDH primers for comparison purposes. Relative mRNA expression was calculated as described by Applied Biosystems (User Bulletin 2: Relative Quantitation of Gene Expression).
Results
Overexpression of Activated Notch Inhibits EC Proliferation
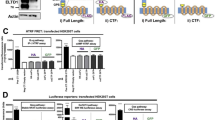
Normal human EC were transduced with the LZRS retroviral expression vector encoding for activated Notch proteins (ie the intracellular domain of the Notch receptor or NIC-1, NIC-2, or NIC-4), and expression was confirmed by Western blot analysis (Figure 1a). Functional activity of the NIC proteins was demonstrated using reporter constructs where luciferase expression was driven by the Hes or Hey-1 gene promoters.23, 25 Cells transduced with the empty vector showed low baseline luciferase levels with the Hey-1-luc reporter construct (Figure 1b). In contrast, cells transduced with NIC-1, NIC-2, or NIC-4 encoding vectors showed a significant increase in luciferase activity compared to the control cells (3–4-fold increase, Figure 1b, P<0.01). Similarly, all three NIC constructs significantly increased luciferase activity in cells transfected with the Hes-1A/B-luc reporter construct (data not shown).
(a) Western blot analysis for expression of activated Notch in transduced EC. Control EC (transduced with the empty LZRS retroviral vector) expressed transmembrane Notch-1 and -2 (TM, arrowheads, approximately 120 kDa) as well as activated NIC-4 (NIC arrow, approximately 75 kDa). In contrast, EC transduced with LZRS-NIC-1 expressed both transmembrane and activated Notch-1 (NIC arrow, 90–110 kDa), but did not express activated Notch-2, which confirmed the specificity of NIC-1 overexpression. Similarly, cells overexpressing NIC-2 possessed both the transmembrane and activated forms of Notch-2, but Notch-1 activation was not detected. As cultured EC constitutively express NIC-4, cells transduced with LZRS-NIC-4 demonstrated an increase in its expression (50% increase on average) with little to no effect on Notch-1 or Notch-2 expression. The results are representative of at least two independent experiments. (b) Induction of Notch transcriptional activity by overexpression of activated Notch. Luciferase activity was measured in EC transduced to express activated Notch and then transfected with a Hey-1-luc reporter construct. The results show significantly higher luciferase activity in NIC-1, NIC-2, and NIC-4-transduced cells compared to vector control cells (3.3-fold, 4.0-fold, and 3.4-fold increase, respectively; P<0.01). Luciferase values were normalized for transfection efficiency. The results are combined data from two experiments performed in triplicate. (c) Notch activation inhibits EC proliferation. Using a WST-1 assay, we found a significant decrease in proliferation of EC overexpressing NIC-1, NIC-2, or NIC-4 compared to the cells transduced with the empty vector control (NIC-1: 2.9-fold decrease; NIC-2: 3.0-fold decrease; NIC-4: 4.3-fold decrease; P<0.01). The results are combined data from three experiments performed in triplicate.
To examine the effect of Notch activation on EC proliferation, cells were transduced with the NIC expression vectors, and WST-1 proliferation assays performed. We have previously demonstrated that over 85% of ECs (range: 87.4–93%) can be reproducibly transduced with LZRS-GFP under our experimental conditions; therefore, the cells were used as a bulk culture and were not selected.21 EC overexpressing NIC-1, NIC-2, or NIC-4 showed a significant reduction in cell proliferation compared to vector-transduced cells (2.9-fold, 3.0-fold, or 4.3-fold decrease, respectively; Figure 1c; P<0.01).
To determine if the decreased proliferation was due to apoptosis or growth arrest, DNA/propidium iodide (DNA/PI) staining was performed to evaluate DNA content. The results revealed relatively little apoptosis in either the vector or NIC-transduced EC, although there was a significant increase in the number of cells in the G0/G1 phase of the cell cycle (Figure 2). The results were confirmed using annexin V staining and flow cytometry which revealed a similar number of EC were annexin V positive under each of the treatment conditions (vector control: 2.3±0.2%, NIC-1: 2.6±0.5%, NIC-2: 1.8±0.3%, NIC-4: 3.8±0.7%). These results confirm and extend previous reports showing NIC-1 or NIC-4 overexpression induce a G0/G1 growth arrest in EC.18, 16
Overexpression of activated Notch results in a G0/G1 arrest, but not apoptosis. DNA/PI staining and flow cytometry revealed a modest, but reproducible increase in the percentage of cells in the G0/G1 phase of the cell cycle in NIC-1, NIC-2, and NIC-4 expressing EC compared to control-transduced cells. The average number of cells in G0/G1 were 73.2±0.4% (vector), 84.9±0.9% (NIC-1, P<0.01), 83.3±1.8% (NIC-2, P<0.01), 85.3±1.5% (NIC-4, P<0.01). However, there was relatively little change in the percentage of cells in apoptosis (sub-G0 DNA content, ranging from 1.9 to 4.3% apoptotic cells). In EC transduced to express Jagged-1, there was no difference in the DNA/PI profiles between EC+Jagged and EC+vector control cells. DNA/PI results are representative of three independent experiments.
Jagged-Induced Notch Activation
Overexpression studies, while informative, can be misleading as protein expression can be substantially different from physiologic conditions. Therefore, experiments were designed using EC transduced with LZRS-Jagged-1 to induce Notch activation in adjacent EC through engagement of the endogenous Notch receptors. Western blot confirmed elevated Jagged-1 expression in the transduced cells, and demonstrated increased levels of transmembrane and activated Notch-1 and Notch-2 as well as activated Notch-4 (Figure 3a). The results were confirmed using the Hes-1A/B luciferase reporter construct, which revealed a three-fold increase in activity in Jagged-1 expressing EC compared to vector control cells (Figure 3b, P<0.01). Under these conditions, Notch activation resulted in a more modest, yet statistically significant, decrease in proliferation (1.3-fold decrease, P<0.01; Figure 3c). No difference was detected in the DNA/PI profiles between LZRS-Jagged-1 and LZRS-vector-transduced cells (Figure 2), which is likely due to the modest changes in proliferation detected in these populations.
(a) Western blot analysis confirmed Jagged-1 overexpression in EC transduced with LZRS-Jagged-1, and demonstrated activation of Notch-1, Notch-2, and Notch-4 in these cells (NIC arrows). Transmembrane expression of Notch-1 and Notch-2 receptors (TM, arrowheads) was also increased in the Jagged-1-transduced EC compared to control cells. Results are representative of at least two experiments. (b) Notch transcriptional activity was examined using the Hes-1A/B-luc reporter construct in EC transduced with Jagged-1 or stimulated with JAG1. Luciferase activity in Jagged-1-transduced EC was 3.0-fold higher than vector control cells (P<0.01). Similarly, EC treated with 40 μM JAG1 peptide had 3.9-fold higher luciferase activity than cells treated with the same concentration of a scrambled control peptide (P<0.01). Luciferase values were normalized for transfection efficiency. Results are combined data from three independent experiments performed in triplicate. (c) WST-1 proliferation assays revealed a modest, yet statistically significant, decrease in proliferation of EC transduced with Jagged-1 compared to vector control cells (1.3-fold decrease, P<0.01). Treatment of cells with JAG1 resulted in a 1.2-fold decrease in proliferation, which was also statistically significant (P<0.05) compared to cells treated with a scrambled control peptide. Results are combined data from at least two experiments performed in triplicate.
To confirm these results, experiments were performed using a previously described Jagged-1 peptide (JAG1) that binds to and activates Notch receptors.26 Initial experiments demonstrated functional activity of the peptides under our experimental conditions. EC treated with 40 μM JAG1 peptide showed, on average, a 3.9-fold increase in Hes-1 transcriptional activity compared to cells treated with a scrambled control peptide in a luciferase reporter assay (Figure 3b, P<0.01). As seen in the Jagged-1 expression studies, JAG1 peptide induced a modest, yet statistically significant reduction, in EC proliferation (1.2-fold decrease, P<0.05; Figure 3c). Taken together, the results indicate Notch activation in proliferating EC induces a G0/G1 growth arrest, but not apoptosis. Decreased proliferation was more dramatic in cells overexpressing activated Notch compared to cells where the endogenous Notch receptors were engaged by the Notch ligand, Jagged-1. This data are consistent with and extends recently published results.16, 17, 18
EC–Cell Contact Induces Hes-1 Expression Via a Notch-Independent Mechanism
Recent studies have suggested Notch activation may be important in the establishment and/or maintenance of EC growth arrest as seen in the quiescent, contact-inhibited EC lining the vasculature in vivo.18 To gain insight into EC contact inhibition and Notch activation, we examined expression of Notch receptors, ligands, and target proteins in actively proliferating vs confluent EC. Total cellular protein was extracted when EC were proliferating (50–60% confluent) or after the cells had reached and maintained confluence for 96 h. There was little to no Notch-1 activation detected in either proliferating or confluent EC, and overall levels of Notch-1 receptor expression were decreased (range: 1.5–2.0-fold) in the confluent cultures (Figure 4a). Expression of activated Notch-4 was slightly, but reproducibly, decreased (range: 1.2–1.5-fold) in confluent EC, while expression of transmembrane Notch-2 receptor was virtually eliminated upon confluence, and there was no evidence of Notch-2 activation in either population (Figure 4a). Similarly, little to no Jagged-1 expression was detected in confluent cells compared to substantial expression in proliferating cells. Dll-1 expression was modest in both populations and did not appear to change. There was also a downregulation of p21, as previously reported.18 These results indicate decreased Notch activation in confluent EC, although it is not completely eliminated due to the lower, but continued expression of NIC-4.
(a) Expression of Notch pathway proteins in proliferating and confluent EC. Western blot analysis of proliferating (Prol) or confluent (Conf) EC proteins from four donors (two are shown) revealed a 1.5–2.0-fold decrease in transmembrane Notch-1 and virtually absent transmembrane Notch-2 expression in confluent cells compared to ample expression in proliferating EC. There was little to no activated Notch-1 or Notch-2 in either cell population. Activated Notch-4 levels were slightly decreased (range: 1.2–1.5-fold) in confluent EC. The Notch ligand Dll-1 was modestly expressed and unchanged between proliferating and confluent cells, while Jagged-1 expression was significantly decreased upon confluency. Hey-1 expression was similar or decreased in confluent cells, while p21 expression was significantly decreased. In contrast, Hes-1 expression was consistently upregulated 1.6–2.0-fold in confluent cells. (b) Luciferase reporter activity in proliferating and confluent EC. There was a two-fold decrease (P<0.05) in Hey-1 promoter driven luciferase activity in confluent EC compared to proliferating cells. In contrast, there was a four-fold increase (P<0.05) in Hes-1 driven activity in confluent vs proliferating EC. (c) γ-secretase inhibition blocked Notch expression and/or activation in proliferating and confluent EC; however, Hes-1 expression remained elevated. Hes-1 expression was similar in confluent EC treated with the γ-secretase inhibitor or treated with DMSO as a control. Hes-1 levels were significantly induced in proliferating cells treated with the inhibitor, although the responsible mechanism has not yet been determined. (d) Induction of Hes-1 in quiescent EC is transcriptionally regulated. Quantitative, real-time RT-PCR was used to measure Hes-1 mRNA expression in proliferating and confluent EC in four different EC donors (two are shown). Hes-1 mRNA levels were normalized to GAPDH expression and the relative quantity of Hes-1 mRNA shown. Although overall levels of Hes-1 mRNA varied between individual EC donors, there was a consistent increase in Hes-1 mRNA in confluent EC compared to proliferating EC (P<0.01).
Despite decreased Notch activation, expression of Hes-1 was upregulated in confluent EC (range: 1.6–2.0-fold, Figure 4a), while Hey-1 expression appeared stable or decreased. To confirm these results, luciferase reporter assays were performed with the Hes-1A/B and Hey1-luc constructs. Cells transfected with an empty luciferase vector as a control possessed low baseline luciferase levels (Figure 4b), while luciferase activity was detectable in proliferating and confluent EC with both reporter constructs. Interestingly, there was a two-fold decrease in Hey-1 promoter driven luciferase activity and a four-fold increase in Hes-1 promoter driven activity in confluent EC compared to proliferating cells (Figure 4b, P<0.05); a finding that is consistent with the Western blot data.
The above results suggest that Hes-1 is being regulated independently of Notch activation in confluent EC, although a modest contribution by activated Notch-4 cannot be dismissed. To confirm our finding, confluent EC were treated with a γ-secretase inhibitor that potently inhibits Notch cleavage and activation.15 As shown in Figure 4c, expression of Notch-1, -2, and -4 were significantly decreased in both proliferating and confluent EC treated with the γ-secretase inhibitor, yet the levels of Hes-1 remained elevated. To determine if Hes-1 expression was being modulated at the transcriptional level, quantitative RT-PCR was used to evaluate Hes-1 mRNA expression in proliferating and confluent EC. Although the levels of Hes-1 mRNA varied between individual EC isolates, there was a consistent increase in Hes-1 mRNA in confluent compared to proliferating EC (Figure 4d, P<0.01). These results not only support the conclusion that Hes-1 is induced in confluent EC, but also indicate that this induction of Hes-1 is being regulated at the transcriptional level.
JNK Signaling Regulates Hes-1 Expression in Confluent EC
To begin investigating the signaling pathways responsible for regulation of Hes-1 expression, confluent EC cultures were treated with pharmacologic agents targeting different signaling proteins (FTI-277 (Ras inhibitor); LY294002 (PI3K inhibitor); SB203580 (p38 inhibitor); PD98059 (MAPK inhibitor); GF109203X (PKC inhibitor); NF-κB activation inhibitor; ERK1/2 activation inhibitor peptide II; U0126 (MEK1/2 inhibitor); JNK inhibitor I) and Hes-1 expression evaluated by Western blot. The results showed that only treatment with JNK inhibitor I (1 μM) significant decreased Hes-1 expression (average decrease: 2.4±0.08-fold), while there was little effect with the other inhibitors (Figure 5a). The results are shown graphically in Figure 5b and suggest that JNK or a downstream target of JNK may be regulating Hes-1 expression in confluent EC. For these studies, each inhibitor was initially tested at 1, 2, and 10X IC50 values (or at concentrations from published work) to determine the appropriate concentration for inhibition of signaling under our experimental conditions (Figure 5c). Figure 5c illustrates the successful blockade of several signaling pathways. In particular, phosphorylated JNK was decreased in EC treated with 1 μM JNK inhibitor 1, although inhibition was incomplete at all of the tested concentrations (Figure 5c). In addition, phosphorylation of ERK1/2, MEK1/2, and p38 (all members of the MAPK superfamily) was blocked demonstrating functional activity of their respective inhibitors (Figure 5c).
(a) Western blot analysis revealed 1 μM JNK inhibitor I (a peptide inhibitor) significantly reduced Hes-1 protein expression in confluent EC, while other inhibitors had little to no effect on Hes-1 expression. The results were confirmed using JNK inhibitor II (a pharmacologic inhibitor, at 3 μM and 10 μM). Representative results from 3 to 4 independent experiments are shown. (b) The fold decrease in Hes-1 expression was calculated from the Western blot experiments, combined, and shown here in graphical form. A value of 1.0 was assigned to DMSO-treated confluent EC (control) and changes in expression calculated. In this figure, negative numbers represent an increase in expression while positive numbers indicate a decrease in expression. Only the inhibition of JNK resulted in a significant decrease in Hes-1 expression (JNK inhibitor I: 2.4±0.08-fold decrease; JNK inhibitor II (3 μM): 1.7±0.01-fold decrease; and JNK inhibitor II (10 μM): 5.6±0.04-fold decrease). (c) Western blot analysis for total and phosphorylated JNK demonstrated that JNK inhibitor I (1 μM) and JNK inhibitor II (3 and 10 μM) blocked phosphorylation. The blockade was not complete at any drug concentration evaluated for JNK inhibitor I, but JNK inhibitor II at the published concentrations of 3 and 10 μM was very effective.27 Phosphorylation of other members of the MAPK superfamily (ERK1/2, MEK1/2, and p38) were successfully blocked by their respective inhibitors under our experimental conditions.
JNK inhibitor I is a cell-permeable, peptide inhibitor consisting of the carboxyl terminal sequence from the JNK binding domain fused with an HIV-TAT sequence to promote entry into the cell. Therefore, we complemented this genetic approach of blocking JNK signaling with a pharmacologic approach using JNK inhibitor II ((SP600125; anthrax(1,9-cd) pyrazol-6 (2H)-one)), an anthrapyrazole. Our initial studies demonstrated that 3 and 10 μM of JNK inhibitor II effectively decreased expression of phosphorylated JNK with more modest decreases noted in total JNK expression (Figure 5c).27 As predicted, treatment with JNK inhibitor II (at both concentrations) significantly decreased Hes-1 expression in confluent EC (average decrease: 1.7±0.01-fold at 3 μM and 5.6±0.04-fold at 10 μM, Figure 5a and b).
Finally, siRNA technology was utilized to confirm JNK signaling was involved in Hes-1 expression in growth arrested EC. Validated siRNA duplexes and a control siRNA were purchased from Qiagen, and their effect on JNK expression in confluent EC measured 48 h after transfection. As shown in Figure 6, JNK expression was significantly inhibited by the JNK, but not control siRNAs (siJNK: average 2.7-fold; siControl: average 1.04-fold decrease). As predicted, Hes-1 protein expression was significantly inhibited by the JNK siRNA (Figure 6, average 6.75-fold decrease) further demonstrating a role for JNK signaling in Hes-1 expression in confluent EC.
siRNA technology was used to block JNK expression in confluent EC. Untreated and control siRNA (siControl) treated confluent EC expressed both JNK and Hes-1 protein; while cells treated with the validated siRNA duplexes showed a significant decrease in both JNK (2.7-fold decrease, on average) and Hes-1 expression (6.75-fold decrease, on average). There was no effect of either siRNA on actin expression although there was a slight decrease in Hes-1 expression (1.26-fold on average) using the control siRNA.
Discussion
Previous studies strongly support a role for the Notch signaling pathway in vasculogenesis; however, much less is known about Notch activation in EC. Recent reports suggest Notch activation induces growth arrest in EC, potentially through mechanisms including p21 repression, inhibition of Rb phosphorylation, and the downregulation of minichromosome maintenance proteins.18, 28 From these studies, it was proposed that Notch induced growth arrest in EC may be linked to the establishment or maintenance of EC quiescence as seen in contact-inhibited EC lining the vasculature. In this report, we set out to examine the effect of Notch activation on normal human EC, and to investigate a possible role for Notch activation in EC quiescence. Our studies confirmed reports that Notch activation induces growth arrest in EC; however, the elevated levels of Hes-1 protein found in quiescent, contact-inhibited EC did not appear to be related to Notch activation, but instead involve signaling through the JNK pathway.
Hes-1 has been identified as a primary target of Notch activation; however, accumulating evidence suggests Hes-1 may also be regulated by Notch-independent mechanisms. For example, mutation of Notch-1 or CBF-1 in mice was shown to alter expression of hes-5, mash-1, and dll-1; however, expression of hes-1 or hes-3 were not altered.29 In addition, Stockhausen et al30 recently showed that transforming growth factor (TGF)-α could induce Hes-1 expression in a neuroblastoma cell line SK-NOBE(2)c, which occurred in the absence of Notch activation.30 The authors demonstrated that Hes-1 expression in this system was dependent on activation of the MAP kinase ERK signaling. Our studies reveal an induction of Hes-1 protein expression (as a result of increased transcription) in quiescent, contact-inhibited EC compared to proliferating cells from the same donor. This expression did not appear to be related to Notch activation, which was decreased in the confluent culture. Furthermore, inhibition of Notch activation with a γ-secretase inhibitor did not alter Hes-1 expression in the quiescent cells. These findings are in contrast to the work of Noseda et al18 who reported that confluent EC activate Notch signaling.18 The difference between these studies may be related to the methods used to evaluate Notch activation. Noseda et al18 examine HRT-1 (an alternative name for Hey-1) target gene expression as a means to monitor Notch activation without evaluating Notch activation directly. Our studies used Western blot analysis for detection of Notch pathway proteins coupled with pharmacologic inhibition of Notch activation, which did not alter Hes-1 expression.
We demonstrate that Hes-1 expression is regulated in confluent EC by the JNK signaling pathway. To our knowledge, this is the first report showing that JNK signaling can regulate Hes-1 expression in a Notch-independent manner. JNK signaling, a subfamily of the MAPK signaling superfamily, is known to regulate the activity of transcription factors including c-Jun, ATF2, ELK-1, p53, and c-Myc. Analysis of the Hes-1 promoter (−954 to +46) indicates that in addition to the previously described binding sites for CBF-1, there are three potential ELK-1 binding sites, a c-jun and ATF2 binding site, as well as a variety of p53 binding sites.31 Several of these sites, including two of the ELK-1 binding sites, are located in regions of the promoter predicted to be important positive regulatory regions.31 Interestingly, these two ELK sites are also within the truncated promoter present in the Hes-1A/B luciferase reporter construct (−194 to +160) that showed significant activity in confluent EC cultures. Further studies are needed to determine if ELK-1 is involved in JNK-mediated Hes-1 expression.
The JNK signaling pathway has been previously linked to the Notch pathway. Studies by Kim et al32 found that Notch activation (whether through ligation of endogenous Notch receptors or overexpression of NIC-1) inhibits JNK signaling.32 They demonstrated direct binding of NIC-1 to JIP-1 (JNK interacting proteins-1), a scaffold protein that normally interacts with MLK3, MKK7 and JNK to facilitate JNK activation. This binding displaced JNK and prevented signaling.32 As confluency results in a decrease in Notch-1 expression in EC and there was no evidence of NIC-1 expression in these cells, JIP-1 should be available to facilitate JNK activation under our experimental conditions. Additional experiments will be needed to determine if this is correct.
References
Radtke F, Raj K . The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer 2003;3:756–767.
Schroeter EH, Kisslinger JA, Kopan R . Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 1998;393:382–386.
Iso T, Kedes L, Hamamori Y . HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 2003;194:237–255.
Iso T, Sartorelli V, Poizat C, et al. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol 2001;21:6080–6089.
Shutter JR, Scully S, Fan W, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev 2000;14:1313–1318.
Leimeister C, Schumacher N, Steidl C, et al. Analysis of HeyL expression in wild-type and Notch pathway mutant mouse embryos. Mech Dev 2000;98:175–178.
Luo B, Aster JC, Hasserjian RP, et al. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol 1997;17:6057–6067.
Uyttendaele H, Marazzi G, Wu G, et al. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 1996;122:2251–2259.
Tsai S, Fero J, Bartelmez S . Mouse Jagged2 is differentially expressed in hematopoietic progenitors and endothelial cells and promotes the survival and proliferation of hematopoietic progenitors by direct cell-to-cell contact. Blood 2000;96:950–957.
Villa N, Walker L, Lindsell CE, et al. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech Dev 2001;108:161–164.
Karsan A . The role of notch in modeling and maintaining the vasculature. Can J Physiol Pharmacol 2005;83:14–23.
Iso T, Hamamori Y, Kedes L . Notch signaling in vascular development. Arterioscler Thromb Vasc Biol 2003;23:543–553.
Uyttendaele H, Ho J, Rossant J, et al. Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium. Proc Natl Acad Sci USA 2001;98:5643–5648.
Clifford GM, Polesel J, Rickenbach M, et al. Cancer risk in the Swiss HIV Cohort Study: associations with immunodeficiency, smoking, and highly active antiretroviral therapy. J Natl Cancer Inst 2005;97:425–432.
Curry CL, Reed LL, Golde TE, et al. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi's sarcoma tumor cells. Oncogene 2005;24:6333–6344.
Liu ZJ, Shirakawa T, Li Y, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol 2003;23:14–25.
Liu ZJ, Xiao M, Balint K, et al. Inhibition of endothelial cell proliferation by Notch1 signaling is mediated by repressing MAPK and PI3K/Akt pathways and requires MAML1. FASEB J 2006;20:1009–1011.
Noseda M, Chang L, McLean G, et al. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol Cell Biol 2004;24:8813–8822.
Foreman KE, Vaporciyan AA, Bonish BK, et al. C5a-induced expression of P-selectin in endothelial cells. J Clin Invest 1994;94:1147–1155.
Kinsella TM, Nolan GP . Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Human Gene Ther 1996;7:1405–1413.
Tang J, Gordon GM, Nickoloff BJ, et al. The helix-loop-helix protein id-1 delays onset of replicative senescence in human endothelial cells. Lab Invest 2002;82:1073–1079.
Tang J, Gordon GM, Muller MG, et al. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces expression of the helix–loop–helix protein Id-1 in human endothelial cells. J Virol 2003;77:5975–5984.
Jarriault S, Brou C, Logeat F, et al. Signalling downstream of activated mammalian Notch. Nature 1995;377:355–358.
Lu C, Gordon GM, Chandran B, et al. Human herpesvirus 8 reactivation and human immunodeficiency virus type 1 gp120. Arch Pathol Lab Med 2002;126:941–946.
Maier MM, Gessler M . Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun 2000;275:652–660.
Nickoloff BJ, Qin JZ, Chaturvedi V, et al. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ 2002;9:842–855.
Miho N, Ishida T, Kuwaba N, et al. Role of the JNK pathway in thrombin-induced ICAM-1 expression in endothelial cells. Cardiovasc Res 2005;68:289–298.
Noseda M, Niessen K, McLean G, et al. Notch-dependent cell cycle arrest is associated with downregulation of minichromosome maintenance proteins. Circ Res 2005;97:102–104.
de la Pompa JL, Wakeham A, Correia KM, et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 1997;124:1139–1148.
Stockhausen MT, Sjolund J, Axelson H . Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res 2005;310:218–228.
Takebayashi K, Sasai Y, Sakai Y, et al. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem 1994;269:5150–5156.
Kim JW, Kim MJ, Kim KJ, et al. Notch interferes with the scaffold function of JNK-interacting protein 1 to inhibit the JNK signaling pathway. Proc Natl Acad Sci USA 2005;102:14308–14313.
Acknowledgements
This work was supported by a grant from the National Institutes of Health CA108450 (KEF) and the American Skin Association (CLC).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Curry, C., Reed, L., Nickoloff, B. et al. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Invest 86, 842–852 (2006). https://doi.org/10.1038/labinvest.3700442
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700442