Abstract
A valuable diagnostic adjunct and important prognostic parameter in alveolar rhabdomyosarcoma (ARMS) is the identification of translocations t(2;13)(q35;q14) and t(1;13)(p36;q14), and the associated PAX3-FKHR and PAX7-FKHR fusion transcripts, respectively. Most RMS fusion gene type studies have been based on reverse transcriptase-polymerase chain reaction (RT-PCR) detection of the fusion transcript, a technique limited by RNA quality and failure of devised primer sets to detect unusual variants. As an alternative approach, we developed a fluorescence in situ hybridization (FISH) assay that can: (1) distinguish between the two most common ARMS-associated fusion genes; (2) identify potential unusual variant translocations; (3) assess histologic components in mixed alveolar/embryonal RMS; and (4) be performed on paraffinized tissue. FISH analyses of 75 specimens (40 ARMS, 16 ERMS, 8 mixed ARMS/ERMS, and 11 non-RMS tumors) using selected cosmid clone, bacterial, P1-derived, and yeast artificial chromosome probe sets were successful in all but two cases. Among specimens with informative results for both FISH and RT-PCR or standard karyotyping, PAX/FKHR classification results were concordant in 94.6% (53/56). The three discordant cases included one exhibiting a t(2;13) by FISH that was subsequently confirmed by repeat RT-PCR, a second showing a rearrangement of the PAX3 locus only (consistent with the presence of a PAX3 variant translocation), and a third revealing a t(2;13) by FISH that lacked this translocation cytogenetically. Both alveolar and embryonal components of the mixed ARMS/ERMS subtype were negative for PAX3, PAX7, and FKHR rearrangements, a surprising finding confirmed by RT-PCR and/or conventional karyotyping. These data demonstrate that FISH with newly designed probe sets is a reliable and highly specific method of detecting t(1;13) and t(2;13) in routinely processed tissue and may be useful in differentiating ARMS from other small round cell tumors. The findings also suggest that FISH may be a more sensitive assay than RT-PCR in some settings, capable of revealing variant translocations.
Similar content being viewed by others
Main
Rhabdomyosarcoma (RMS) is the most common pediatric soft-tissue sarcoma related to the myogenic lineage. RMS can be divided into three primary histologic subtypes including embryonal RMS (ERMS), alveolar RMS (ARMS), and pleomorphic (anaplastic) RMS.1, 2 Anaplasia can be identified in any subtype of RMS.2 ERMS encompasses the botryoid and spindle cell variants. ARMS may present as a solid pattern or mixed with both embryonal and alveolar features. These histologic subtypes are one of the most useful prognostic parameters in RMS.1, 3
The majority (80%) of ARMS are associated with specific chromosomal translocations occurring between chromosomes 2 and 13 or less commonly 1 and 13 leading to the fusion of either the transcription factor PAX3 in the former case or the related transcription factor PAX7 in the latter case to another transcription factor FKHR.4, 5 The remaining 20% of ARMS are translocation or fusion-negative forming a basically unexplored group, although two variant translocation associated fusion transcripts (PAX3-AFX and PAX3-NCOA1) have been described.6, 7
Although the different PAX-FKHR fusion genes are readily identified by reverse transcriptase-polymerase chain reaction (RT-PCR), this technique may be limited by RNA quality and failure of devised primer sets to detect unusual variants. Previous FISH studies have been performed on a small series of RMS samples.8, 9 Therefore, we wished to develop a novel FISH assay that can distinguish between the two most common ARMS-associated fusion genes, identify potential unusual variant translocations, and provide reliable performance on formalin-fixed, paraffin-embedded tissue. We also assessed both alveolar and embryonal components of the mixed histologic subtype by FISH using paraffin-embedded tissue sections in which morphologic features could be identified.
Materials and methods
Histopathologic Assessment
The histopathologic classification of the RMS specimens was confirmed by the Intergroup Rhabdomyosarcoma Study Group (IRSG) Pathology Review Committee. Tumors were assessed in accordance with the International Classification of Rhabdomyosarcoma (ICR) criteria.3 All of the mixed ARMS/ERMS cases had well-characterized alveolar and embryonal components (Figure 1); equivocal cases were omitted.
Tumor Samples
In total, 40 ARMS samples including 23 fusion-negative cases selected from the IRSG database, 16 ERMS samples, and eight mixed ARMS/ERMS samples were analyzed. In addition, 11 non-RMS tumors (three Ewing sarcomas, three neuroblastomas, two inflammatory myofibroblastic tumors, and three leiomyosarcomas) were included in this study. The histopathologic types of the samples studied are listed in Table 1.
Cytogenetic Analysis
Standard culture and harvesting procedures were performed, as described previously.10 Metaphase cells were banded with Giemsa trypsin, and the karyotypes were expressed according to the International System for Human Cytogenetic Nomenclature 1995.11
RT-PCR Analysis
RT-PCR analysis for the presence of the PAX3-FKHR or PAX7-FKHR fusion transcript was performed as previously described.12
Probe Design and Development
Bacterial artificial chromosome (BAC), P1-derived artificial chromosome (PAC), yeast artificial chromosome (YAC) and cosmid clones for the PAX3, PAX7, and FKHR gene regions were identified utilizing the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview), the Ensembl Genome Browser (http://www.ensembl.org), and the Whitehead Institute/MIT Center Genome Browser (http://www-genome.wi.mit.edu/cgi-bin/contig/yac_info). The probe mixtures used are listed in Table 2. Combinations of probe sets were fashioned to flank and span each gene.
Fluorescence In Situ Hybridization
Two-color FISH studies were performed on cytologic touch preparations (n=25), in situ sections (n=2), and formalin-fixed, paraffin-embedded tissue sections (n=48). Probes were directly labeled by nick translation with either Spectrum Green or Spectrum Orange-dUTP utilizing a modification of the manufacturer's protocol (Vysis, Downers Grove, IL, USA). An amount of 1 μg of DNA for each of three probes, 1.5 μg of DNA for each of two probes, or 3 μg of DNA for one probe was combined to total 3 μg of DNA per label. All nick translation reagents were then multiplied by the total μg of DNA used in the cocktail. Amounts ranging from 200 to 600 ng of each probe were hybridized to the target DNA and blocked approximately 15 times with a combination of Human Cot-1 DNA (Invitrogen, Carlsbad, CA, USA) and human placental DNA.
Prior to hybridization, the touch preparations and in situ slides were pretreated in 2 × saline sodium citrate (SSC) at 72°C for 2 min and in pepsin solution (20 μl 10% pepsin in 50 ml of 0.1 N hydrochloric acid (HCl)) at 37°C for 3 min, washed in 1 × phosphate-buffered saline (PBS) at room temperature for 5 min, fixed in 1% formaldehyde at 4°C for 5 min, and again washed in 1 × PBS at room temperature for 5 min. The slides were then dehydrated in an ethanol series (70, 85, and 100%) at room temperature for 2 min each and air-dried. The cells and probes were codenatured at 75°C for 1 min and incubated at 37°C overnight using the HYBrite™ denaturation/hybridization system (Vysis). Formalin-fixed, paraffin-embedded tissue sections were cut at a thickness of 4 μm. Tissue sections were deparaffinized in Hemo-De three times at room temperature for 10 min each, followed by dehydration in 100% ethanol twice for 5 min each, and air-dried. Tissue sections were then pretreated in 0.2 N HCl at room temperature for 20 min and in 1 M sodium thiocyanate at 80°C for 30 min, rinsed in distilled water at room temperature for 1 min, followed by washing in 2 × SSC twice for 5 min each, digested in pepsin solution (2.5 mg/ml in 0.9% sodium chloride, pH 2.0) at 37°C for 10 min, and finally washed in 2 × SSC twice at room temperature for 5 min each. The cells and probes were codenatured at 74°C for 6 min and incubated at 37°C overnight using the HYBrite™ denaturation/hybridization system.
Posthybridization washing was performed in 0.4 × SSC/0.3% NP-40 at 72°C for 2–3 min, followed by 2 × SSC/0.1% NP-40 at room temperature for 1–2 min. The slides were air-dried in the dark and counterstained with 4′,6-diamidino-2-phenylindole (DAPI II; Vysis).
Before reviewing the FISH assay, we confirmed the appropriate areas of both alveolar and embryonal components of the mixed histologic subtype using a parallel hematoxylin and eosin (H&E) stained section. Hybridization signals were assessed in 200 interphase nuclei with strong, well-delineated signals and distinct nuclear borders by two different individuals. An interphase cell specimen was interpreted as abnormal if spanning probe signals for the PAX3 or PAX7 genes fused with the spanning probe signal for the FKHR gene, or if a split of flanking probe signals was detected in more than 10% of the cells evaluated (more than two standard deviations above the average false-positive rate). Positive controls included one t(1;13) positive (CW9019) and two t(2;13) positive (RH28 and RH30) ARMS cell lines. Negative controls included normal peripheral blood lymphocytes, cytologic touch preparations and paraffin-embedded tissue sections of pathologically unremarkable skeletal muscle, and one translocation negative ERMS cell line RD. Images were acquired by use of the CytoVision Image Analysis System (Applied Imaging, Santa Clara, CA, USA).
Results
In the present study, cytologic touch preparations, in situ sections, or formalin-fixed, paraffin-embedded tissue sections from 75 tumor specimens were evaluated using the FISH technique. The cytogenetic, RT-PCR and FISH findings of the 64 RMS specimens are summarized in Table 1. The 11 non-RMS tumors were normal by FISH. FISH studies were successfully performed on all but two RMS cases. Prolonged exposure to formalin may be responsible for the two failed hybridizations despite repeated efforts.
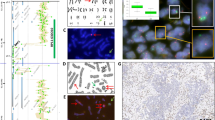
Among all specimens with informative results for both FISH and RT-PCR or conventional cytogenetic analysis, PAX/FKHR classification results (ie positive or negative) were concordant in 94.6% (53/56). When normal metaphase cells are obtained, they are assumed to arise from normal stromal cells, and conventional cytogenetic studies are therefore considered uninformative. There were two discordant cases between FISH and RT-PCR and one discordant case between FISH and conventional cytogenetic analysis (Table 1). One discordant case exhibited rearrangements and fusion of the PAX3 and FKHR gene loci by FISH (Figure 2), but had been initially reported as negative by RT-PCR from the IRSG database. Repeat RT-PCR analysis confirmed the presence of a PAX3-FKHR fusion transcript. A second discordant case exhibited splitting of the PAX3 breakpoint flanking probe set and amplification of the probe signal located just distal to the PAX3 breakpoint (Figure 3a). However, FISH analysis with the FKHR breakpoint flanking probe set revealed normal results; the signals remain juxtaposed to each other (Figure 3b). FISH analysis with the PAX3 and FKHR breakpoint spanning probes showed amplification of the PAX3 spanning probe, but no fusion with or amplification of the FKHR spanning probe (Figure 3c). These findings suggest the presence of a possible PAX3 variant translocation. In the third discordant case, PAX3 and FKHR rearrangements and PAX3-FKHR fusion were detected by FISH (Figure 4a), but cytogenetic analysis showed no clear evidence of the t(2;13) translocation (Figure 4b). Unfortunately, we were unable to evaluate this case by RT-PCR because the quality of RNA was suboptimal.
FISH analyses performed on cytologic touch preparations of Case 3 (ARMS). (a) FISH analysis with the FKHR breakpoint flanking probe set (proximal portion, orange; distal portion, green) shows splitting of the orange and green signals, indicating disruption of FKHR. (b) FISH analysis with the PAX3 breakpoint flanking probe set (proximal portion, orange; distal portion, green) shows splitting of the orange and green signals, indicating disruption of PAX3. (c) FISH analysis with the PAX3 (orange) and FKHR (green) spanning probes shows juxtaposed green and orange or overlapping yellow-white signals (arrows), confirming the presence of a PAX3-FKHR fusion.
FISH analyses performed on cytologic touch preparations of Case 13 (ARMS). (a) FISH analysis with the PAX3 breakpoint flanking probe set (proximal portion, orange; distal portion, green) reveals splitting of the orange and green signals and amplification of the green signal (located just distal to the PAX3 breakpoint), indicating disruption and amplification of PAX3. (b) FISH analysis with the FKHR breakpoint flanking probe set (proximal portion, orange; distal portion, green) reveals normal results; the orange and green signals remain juxtaposed to each other. (c) FISH analysis with the PAX3 (orange) and FKHR (green) spanning probes reveals amplification of the orange signal, but no fusion with or amplification of the green signal. These findings suggest the presence of a possible PAX3 variant translocation. (A single normal cell is present in lower right-hand corner.)
FISH and G-banding analyses of Case 34 (solid variant ARMS). (a) FISH analysis with the PAX3 (orange) and FKHR (green) spanning probes reveals PAX3-FKHR fusion signals (arrows), indicating the presence of a t(2;13) translocation. (b) Representative karyotype showing the following abnormal complement: 46,XY,del(6)(p21p25). Arrows indicate the involved breakpoints on the normal chromosome 6 homologue on the left. Cytogenetic studies did not reveal the characteristic translocation involving chromosomes 2 and 13.
In contrast to the alveolar histologic subtype, all cases of ERMS and non-RMS tumors were negative for PAX3, PAX7, and FKHR gene loci rearrangements. Moreover, both alveolar and embryonal components of the mixed ARMS/ERMS were negative for PAX and FKHR rearrangements (Figure 5), a finding confirmed by RT-PCR or conventional karyotyping.
FISH analysis performed on paraffin-embedded tissue sections of Case 57 (mixed ARMS/ERMS) using the FKHR breakpoint flanking probe set (proximal portion, orange; distal portion, green). The orange and green signals remain fused in both embryonal (a) and alveolar (b) components of the mixed histologic subtype, indicating no rearrangement of FKHR.
Discussion
The diagnosis of pediatric small round cell tumors can be difficult because of overlapping histologic and/or immunohistochemical features. Moreover, the increasing use of minimal biopsy makes the diagnosis of these tumors more challenging. The ability to identify tumor-specific chromosomal translocations and associated fusion gene transcripts using interphase FISH and/or RT-PCR on paraffin-embedded material can be extremely useful in such cases.13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23
In the present study, we describe the largest series of the detection of RMS fusion gene type by FISH to date. Previous FISH studies have been applied to relatively few ARMS cases exhibiting a t(2;13) translocation.8, 9 Moreover, most of these have been performed on touch preparations of fresh or snap-frozen tissue, or on metaphase spreads and/or interphase nuclei from tumor cells grown in short-term culture. However, fresh or frozen tissue for analysis is not always available, and the most widely available tumor tissue is formalin-fixed and paraffin-embedded. Here, we have developed an optimized two-color FISH assay, which is applicable to formalin-fixed, paraffin-embedded tissue and allows a more extensive study of archival material from patients for whom there are detailed data of treatment and outcome.
The newly developed probes that we have described localize to the breakpoints on chromosomes 1, 2, and 13 and can distinguish between the t(2;13) and t(1;13) translocations. The distinction between these translocations in ARMS is important, because patients with t(2;13) have a more adverse outcome than patients with t(1;13) despite indistinguishable histologic features.5 On the other hand, previous FISH studies have used only cosmid probes to detect the disruption of FKHR gene locus at 13q14.8, 9 In the current study, we used BAC clones covering the entire FKHR gene in a single hybridization experiment. In contrast to the cosmid probes, the signals generated by the BAC probes are larger and brighter. This is particularly important for working with paraffin-embedded tissue in the routine clinical diagnostic setting and revealing chromosomal rearrangements in cases with complex variant translocations.
Although there was a high correlation between the results of FISH and those of RT-PCR or conventional cytogenetic analysis, three discordant cases were identified. One case (Case 3), while positive for the presence of the t(2;13) translocation by FISH, did not show a PAX3-FKHR fusion transcript by the initial RT-PCR. Repeat RT-PCR identified the presence of PAX3-FKHR fusion transcript. This finding indicates that these approaches are complementary.
An additional advantage of FISH is its ability to identify unusual variant or cryptic translocations. Two PAX3 variant translocations have been previously described: PAX3-AFX and PAX3-NCOA1.6, 7 Our FISH studies suggest that one of the discordant cases, Case 13, also represents a PAX3 variant translocation. FISH analysis of this case revealed rearrangement of the PAX3 gene with no fusion of signals occurring between the PAX3 and FKHR gene loci. We are currently performing rapid amplification of cDNA ends experiments in an effort to further characterize this rearrangement.
In the third discordant case (Case 34), the t(2;13) or t(1;13) translocations or derivative chromosomes 1, 2, or 13 originating from them were not observed on conventional cytogenetic analysis, but FISH analysis revealed the presence of the t(2;13) translocation. Similarly, several cases of ARMS have previously been reported that failed to demonstrate tumor-specific chromosomal translocations on cytogenetic analysis, but were shown to express PAX-FKHR fusion transcripts by RT-PCR and/or FISH.24, 25, 26 Possible explanations for these findings are the difficulty of identifying the small derivative chromosome 13 or the presence of a cryptic translocation. FISH and RT-PCR analyses are valuable techniques that should be performed to detect the presence of tumor-specific chromosomal translocations or associated fusion gene transcripts even when cytogenetic analysis is successful.
Some RMS cases exhibit a mixture of alveolar and embryonal patterns, and these have been called mixed alveolar/ERMS. According to the ICR criteria, tumors with any alveolar features are classified as ARMS.3 In our FISH analysis, both the alveolar and embryonal components of the mixed histologic subtype were negative for PAX3, PAX7, and FKHR rearrangements. These surprising results were confirmed by RT-PCR or conventional karyotyping in all cases that sufficient material was available for analysis. To the contrary, Biegel et al27 reported a case of mixed ARMS/ERMS with a t(1;13)(p36;q14) translocation. The reason for this discrepancy is unclear. Of note, in two cases of mixed ARMS/ERMS, N-myc amplification was present only in the alveolar component.28 Additional studies are required to determine the genetic characteristics of this histologic subtype.
In summary, these findings: (1) demonstrate that FISH analysis with these newly designed probe sets is a reliable and highly specific method of detecting t(1;13) and t(2;13) in routinely processed tissue (including formalin-fixed, paraffin-embedded tissue) and may be useful in differentiating ARMS from other small round cell tumors; (2) suggest that FISH may be a more sensitive assay than RT-PCR in some settings (capable of revealing variant translocations); and show that both the alveolar and embryonal components of the mixed ARMS/ERMS are negative for PAX3, PAX7, and FKHR gene loci rearrangements.
References
Parham DM, Barr FG . Embryonal rhabdomyosarcoma. In: Fletcher CDM, Unni KK, Mertens F (eds). WHO Classification of Tumours, Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press: Lyon, France, 2002, pp 146–154.
Kodet R, Newton Jr WA, Hamoudi AB, et al. Childhood rhabdomyosarcoma with anaplastic (pleomorphic) features: a report of the Intergroup Rhabdomyosarcoma Study. Am J Surg Pathol 1993;17:443–453.
Newton Jr WA, Gehan EA, Webber BL, et al. Classification of rhabdomyosarcomas and related sarcomas: pathologic aspects and proposal for a new classification – an Intergroup Rhabdomyosarcoma Study. Cancer 1995;76:1073–1085.
Barr FG . Molecular genetics and pathogenesis of rhabdomyosarcoma. J Pediatr Hematol Oncol 1997;19:483–491.
Sorensen PHB, Lynch JC, Qualman SJ, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the Children's Oncology Group. J Clin Oncol 2002;20:2672–2679.
Barr FG, Qualman SJ, Macris MH, et al. Genetic heterogeneity in the alveolar rhabdomyosarcoma subset without typical gene fusions. Cancer Res 2002;62:4704–4710.
Wachtel M, Dettling M, Koscielniak E, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res 2004;64:5539–5545.
Biegel JA, Nycum LM, Valentine V, et al. Detection of the t(2;13)(q35;q14) and PAX3-FKHR fusion in alveolar rhabdomyosarcoma by fluorescence in situ hybridization. Genes Chromosomes Cancer 1995;12:186–192.
McManus AP, O'Reilly MAJ, Pritchard-Jones K, et al. Interphase fluorescence in situ hybridization detection of t(2;13)(q35;q14) in alveolar rhabdomyosarcoma: a diagnostic tool in minimally invasive biopsies. J Pathol 1996;178:410–414.
Bridge JA, Liu J, Qualman SJ, et al. Genomic gains and losses are similar in genetic and histologic subtypes of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer 2002;33:310–321.
Mitelman F (ed). An International System for Human Cytogenetic Nomenclature. S Karger: Basel, 1995.
Barr FG, Xiong QB, Kelly K . A consensus polymerase chain reaction-oligonucleotide hybridization approach for the detection of chromosomal translocations in pediatric bone and soft tissue sarcomas. Am J Clin Pathol 1995;104:627–633.
Reichmuth C, Markus MA, Hillemanns M, et al. The diagnostic potential of the chromosome translocation t(2;13) in rhabdomyosarcoma: a PCR study of fresh-frozen and paraffin-embedded tumour samples. J Pathol 1996;180:50–57.
Anderson J, Renshaw J, McManus A, et al. Amplification of the t(2;13) and t(1;13) translocations of alveolar rhabdomyosarcoma in small formalin-fixed biopsies using a modified reverse transcriptase polymerase chain reaction. Am J Pathol 1997;150:477–482.
Nagao K, Ito H, Yoshida H, et al. Chromosomal rearrangement t(11;22) in extraskeletal Ewing's sarcoma and primitive neuroectodermal tumour analysed by fluorescence in situ hybridization using paraffin-embedded tissue. J Pathol 1997;181:62–66.
Kumar S, Pack S, Kumar D, et al. Detection of EWS-FLI-1 fusion in Ewing's sarcoma/peripheral primitive neuroectodermal tumor by fluorescence in situ hybridization using formalin-fixed paraffin embedded tissue. Hum Pathol 1999;30:324–330.
Qian X, Jin L, Shearer BM, et al. Molecular diagnosis of Ewing's sarcoma/primitive neuroectodermal tumor in formalin-fixed paraffin-embedded tissues by RT-PCR and fluorescence in situ hybridization. Diagn Mol Pathol 2005;14:23–28.
Lee W, Han K, Harris CP, et al. Use of FISH to detect chromosomal translocations and deletions: analysis of chromosome rearrangement in synovial sarcoma cells from paraffin-embedded specimens. Am J Pathol 1993;143:15–19.
Nagao K, Ito H, Yoshida H . Chromosomal translocation t(X;18) in human synovial sarcomas analyzed by fluorescence in situ hybridization using paraffin-embedded tissue. Am J Pathol 1996;148:601–609.
Yang P, Hirose T, Hasegawa T, et al. Dual-colour fluorescence in situ hybridization analysis of synovial sarcoma. J Pathol 1998;184:7–13.
Tsuji S, Hisaoka M, Morimitsu Y, et al. Detection of SYT-SSX fusion transcripts in synovial sarcoma by reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Am J Pathol 1998;153:1807–1812.
Lu YJ, Birdsall S, Summersgill B, et al. Dual colour fluorescence in situ hybridization to paraffin-embedded samples to deduce the presence of the der(X)t(X;18)(p11.2;q11.2) and involvement of either the SSX1 or SSX2 gene: a diagnostic and prognostic aid for synovial sarcoma. J Pathol 1999;187:490–496.
Fritsch MK, Bridge JA, Schuster AE, et al. Performance characteristics of a reverse transcriptase-polymerase chain reaction assay for the detection of tumor-specific fusion transcripts from archival tissue. Pediatr Dev Pathol 2002;6:43–53.
Barr FG, Chatten J, D'Cruz CM, et al. Molecular assays for chromosomal translocations in the diagnosis of pediatric soft tissue sarcomas. JAMA 1995;273:553–557.
Arden KC, Anderson MJ, Finckenstein FG, et al. Detection of the t(2;13) chromosomal translocation in alveolar rhabdomyosarcoma using the reverse transcriptase-polymerase chain reaction. Genes Chromosomes Cancer 1996;16:254–260.
Frascella E, Lenzini E, Schafer BW, et al. Concomitant amplification and expression of PAX7-FKHR and MYCN in a human rhabdomyosarcoma cell line carrying a cryptic t(1;13)(p36;q14). Cancer Genet Cytogenet 2000;121:139–145.
Biegel JA, Meek RS, Parmiter AH, et al. Chromosomal translocation t(1;13)(p36;q14) in a case of rhabdomyosarcoma. Genes Chromosomes Cancer 1991;3:483–484.
Hachitanda Y, Toyoshima S, Akazawa K, et al. N-myc gene amplification in rhabdomyosarcoma detected by fluorescence in situ hybridization: its correlation with histologic features. Mod Pathol 1998;11:1222–1227.
Acknowledgements
We thank Ms Kimberly Christian for her expert secretarial assistance and Dr Thomas Seemayer for his valuable comments. This work was supported in part by NIH (CA89461, CA36727, and CA98543) and the Gladys Pearson Fellowship in Pediatric Cancer and Genetics.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nishio, J., Althof, P., Bailey, J. et al. Use of a novel FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of alveolar rhabdomyosarcoma. Lab Invest 86, 547–556 (2006). https://doi.org/10.1038/labinvest.3700416
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700416
Keywords
This article is cited by
-
Immunohistochemical detection of PAX-FOXO1 fusion proteins in alveolar rhabdomyosarcoma using breakpoint specific monoclonal antibodies
Modern Pathology (2021)
-
Rhabdomyosarcoma
Nature Reviews Disease Primers (2019)
-
Paratesticular alveolar rhabdomyosarcomas do not harbor typical translocations: a distinct entity with favorable prognosis?
Virchows Archiv (2018)
-
Fibroma of tendon sheath around large joints: clinical characteristics and literature review
BMC Musculoskeletal Disorders (2017)
-
The comparative utility of fluorescence in situ hybridization and reverse transcription-polymerase chain reaction in the diagnosis of alveolar rhabdomyosarcoma
Virchows Archiv (2015)