Abstract
Histiocytic necrotizing lymphadenopathy (HNL), a disease of unknown cause, is characterized pathologically by the presence of plasmacytoid dendritic cells (pDCs), which are frequently mixed with oligoclonal T cells (OTCs) and myeloid cells. Toll-like receptors (TLRs 1–10) are a family of pattern recognition receptors of DCs. To investigate the interactions between pDCs and T cells, and to look for an etiology of HNL, we studied 24 HNLs for the profile of TLRs. Transcripts of TLR7, a receptor on pDCs for single-stranded RNA, were found in every case, confirming the universal presence of pDCs. Transcripts of TLR9, another receptor on pDCs for microbial unmethylated CpG-rich DNA, were correlated with OTCs, implying T-cell expansion stimulated by TLR9+ pDCs in response to a microbe. Because PCRs for bacterial 16S rDNAs were negative in the lymph nodes, a bacterial origin seems unlikely, but a virus remains a possible candidate. The pDCs lacked the maturation marker CD83, which suggested ineffective stimulation of T cells and might account for the usually benign course of HNL. Taken together, these data illustrate a novel approach, based upon TLR transcript analysis, for the integration of pathology, immunology, and clinical findings of HNL.
Similar content being viewed by others
Main
Histiocytic necrotizing lymphadenopathy (HNL), also known as Kikuchi's disease, is a self-limiting lymphadenopathy with characteristic clinical and histopathologic features.1, 2 The disease is especially common in young Asian females, and usually involves cervical lymph nodes with patchy subcortical necrosis and abundant nuclear debris surrounded by plasmacytoid dendritic cells (pDCs) and cytotoxic T cells.3 Recently, myeloperoxidase-positive histiocytes were reported in HNL, suggesting also the participation of monocytes or myeloid DCs (mDCs).4 It is noteworthy that the T cells were frequently oligoclonal, implying a specific immune response in the pathogenesis of HNL.5, 6 Nonetheless, details of interactions between DCs and T cells, and the significance of myeloid vs lymphoid DCs, are poorly understood, partially because the etiology of HNL is still unknown.
DCs are migratory antigen-presenting cells derived from hematopoietic stem cells, and include both the better-characterized mDCs (CD11c+) and the still-enigmatic lymphoid or pDCs.7 Immature DCs in the peripheral tissue may capture foreign antigens through pattern recognition receptors, such as the family of Toll-like receptors (TLRs 1–10).8, 9 Stimulated by the foreign antigens, immature DCs migrate towards lymph nodes, develop into mature DCs, and present antigens to T lymphocytes to initiate a primary immune response.10, 11
pDCs were originally identified in the T zone of reactive lymph nodes, based on their plasma-cell-like morphology.12 They have a typical immunophenotype, being CD4+, CD11c−, CD36+, CD45RA+, CD68+, CD74+, and CD123+. They were negative for the myeloid markers CD14 and CD16, the T-cell marker CD3, and the B-cell marker CD19. Because of the lack of lineage-specific markers, the origin of pDCs has been controversial,13, 14, 15 but the culture of pDCs from lymphoid precursors suggested that pDCs were identical to lymphoid DCs.16
TLRs are pattern recognition receptors of the innate immune system.17, 18 There are currently 10 TLRs, widely present in a variety of cells. TLR7 and TLR9 have been found mainly on pDCs, but mDCs express a broad spectrum of TLRs.19, 20, 21, 22 The ligands for some TLRs are known.23 For example, the TLR2/TLR1 or TLR2/TLR6 complex recognizes lipoproteins of Gram-positive bacteria. TLR3 recognizes double-stranded RNAs. TLR4 recognizes lipopolysaccharides of Gram-negative bacteria. TLR5 recognizes bacterial flagellin. TLR9 recognizes unmethylated G–C-rich DNA of bacterial or viral origin. The natural ligand for TLR7 is single-stranded RNA,24, 25 but some antiviral nucleotide analogues may also bind TLR7. The ligands for TLR8 and TLR10 remain unidentified. In concordance with their recognition functions, patterns of TLR expression have clinical implications.26, 27 Consequently, a detailed analysis of the TLR repertoire in HNL, as receptors for foreign antigens, might give clues to the etiology or histopathogenesis of HNL.
TLRs may also function as costimulatory receptors,28 which was well established for CD4+ helper T cells but remained controversial for CD8+ cytotoxic T cells until recently.29, 30, 31 The costimulatory action has direct clinical implications too. For example, modulation of tissue TLR2 and TLR4 during the early phases of polymicrobial sepsis correlates with mortality.32 These data suggest that a detailed analysis of the TLR repertoire in HNL might also provide evidence for the interaction between DCs and cytotoxic T cells, and might predict the clinical outcome for HNL.
In this report, we describe an RT-PCR approach for characterizing the TLR repertoire. By correlating the TLR repertoire with either the clonality of T cells or the clinical manifestation, we expect to find a distinct TLR pattern that might shed light on the possible cause of HNL, clarify the details of DC–T-cell interaction, and predict the clinical outcome.
Materials and methods
Tissue Samples
We collected 24 cases of HNL. Diagnoses were made on cervical lymph node biopsies at the time of initial presentation in the Pathology Department of the National Taiwan University Hospital between 1998 and 2001. All biopsies showed characteristic patchy subcortical necrosis and nuclear debris, with prominent infiltration of pDCs and T cells, but no granulocytes. Clinical data were obtained from the medical records. Patients with serological or cultural evidence of bacterial or viral infections were excluded. Patients suspected of autoimmune disease, such as SLE, were excluded, based on clinical presentation, presence of antinuclear antibodies, and a protracted course with multiple system involvement. A summary of the clinical data for HNL patients is given in Table 1. All patients were Taiwanese. They usually had cervical lymphadenopathy without systemic manifestations except for fever. All of the patients with HNL had a benign course, with spontaneous resolution of the lymphadenopathy within 6 months.5
T-Cell-Receptor (TCR) γ Rearrangement
TCR-γ rearrangement was determined according to a protocol developed in our laboratory.5 Briefly, four separate nested PCRs were used for examining the four variable regions of TCR-γ, VγI, VγII, VγIII, and VγIV. The primers were
VI, 5′-TCAGGAATCAGTCCAGGAAAGTAT-3′;VII, 5′-GAAAGGAATCTGGCATTCCG-3′;VIII, 5′-AAGCAACAAAGTGGAGGCAAGAAAG-3′;VIV, 5′-CTCACACTC(T/C)CACTTC-3′;Jγ1/2, 5′-CAAGTGTTGTTCCACTGCC-3′;Jγp, 5′-TTGTTCCGGGACCAAATACC-3′;Jγp1/2, 5′-GTTACTATGAGC(C/T)TAGTC-3′;V1′, 5′-6-FAM-TCTGG(A/G)GTCTATTACTGTGC-3′;VII′, 5′-6-FAM-ATAGCTACCTACTACTGTGC-3′;VIII′, 5′-6-FAM-ATGGCCGTTTACTACTGTGC-3′;VIV′, 5′-6-FAM-GAGGTGGTGTACCACTGTGC-3′;Jγ1/2′, 5′-AGTGTTGTTCCACTGCCAAAGAGTTT-3′;Jγp′, 5′-AGCTTTGTTCCGGGACCAAATACCTT-3′;Jγp1′, 5′-AGCTTAGTCCCTTCAGCAAATATCTT-3′;Jγp2′, 5′-AGCCTAGTCCCTTTTGCAAACGTCTT-3′.
For example, to assay VγI-J recombination, we used a VγI-specific primer (V1) and a mixture of J-primers (Jγ1/2, Jγp, and Jγp1/2) to amplify 0.1 μg of extracted DNA in an initial PCR. This was followed by a nested PCR, with use of a second fluorescence-labeled VγI-specific primer (V1′), a second mixture of J-primers (Jγ1/2′, Jγp′, Jγp1′, and Jγp2′), and 2 μl of the initial PCR mixture as the DNA template. Illustrative examples of polyclonal and oligoiclonal TCR rearrangements are shown in Figure 1.
Analysis of Toll-Like Receptor Transcripts by RT-PCR
The sequences of the primers for RT-PCR are listed in Table 2, which also gives the GenBank accession number of each TLR, the positions of the primers, and the sizes of the PCR products. The sequences and genomic structures of TLRs are also available in references Barton and Medzhitov23, Wiemann et al,33 Rock et al,34 Medzhitov et al,35 Chaudhary et al,36 Takeuchi et al,37 Du et al38 and Chuang and Ulevitc.39 For example, to evaluate the TLR1 transcripts, a forward primer, 5′-ATAACAAAGGCATATTGGGCA-3′ (174–194 in exon 3), and an antisense primer, 5′-RS-TGTTCTTCAGATCATCTTGAT-3′ (241–221 in exon 4), were used. The ‘RS’ was a random sequence, 5′-TGACAAACTGTGTTCACTAGC-3′, for increased PCR specificity and incorporation of fluorescent labels.
Reverse transcription was generally done in a 20-μl mixture at 40°C for 50 min. The mixture included 0.1 μg of RNA purified from formalin-fixed, paraffin-embedded tissue blocks, 0.5 μM antisense primer, 200 U of reverse transcriptase, 50 mM Tris-HCl at pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, and 200 μM of each dNTP. The cDNA from 2 μl of the reverse transcription mixture was amplified by a standard PCR, with the forward primer and a fluorescence-labeled primer of the random sequence, 5′-TET-CTTCTGACAAACTGTGTTCACTAGC-3′. The standard PCR was a 20 μl PCR mixture with the DNA template, 0.3 μM of each primer, 15 mM Tris-HCl at pH 8.0, 1.5 mM MgCl2, 50 mM KCl, 200 μM of each dNTP, and 1 U of Taq polymerase. The reaction mixture was subjected to 35 cycles of PCR. Each cycle consisted of denaturation at 94°C for 45 s, annealing at 45°C for 45 s, and extension at 72°C for 45 s.
At the end of 35 cycles, a portion of the PCR product was loaded on and separated by a high-resolution electrophoretic system (ABI377 with GeneScan software, Perkin-Elmer, Foster City, CA, USA). The sizes for the RT-PCR products of TLRs are listed in Table 2. A reaction for β2M was run as a positive control, and a reaction under the same conditions except for the omission of reverse transcriptase was run as a negative control.
A representative result is shown in Figure 2. A TLR is deemed positive if its peak is greater than 10% of β2M.
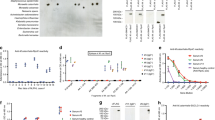
Representative GeneScan tracings for TLR repertoire. Three cases are shown in the upper (a), middle (b), and lower (c) horizontal panels. From left to right, the four vertical columns show β2M (as internal positive control), TLR1, TLR7, and TLR9. All three cases were TLR1+ and TLR7+, but TLR9 varied from being weakly positive (upper panel) to negative (middle panel) to strongly positive (lower panel).
Correlation between TLR Repertoire and TCR
The 24 cases of HNL were placed in an oligoclonal group and a polyclonal group. Two-sample comparison was done based on a binomial proportion test for ratios and a Mann–Whitney test for continuous data. The results are summarized in Figure 3 and Table 3. For confirmation, a stepwise regression analysis based on the logistic model was carried out at the Biostatistics Center at the National Taiwan University Hospital.
Distribution of TLR transcripts in polyclonal and oligoclonal HNLs. The polyclonal group (left) and the oligoclonal group (right) are shown side by side for comparison. The distribution of TLR transcripts is summarized in the box–whisker plot. The box shows the 25th percentile, the median (the 50th percentile), and the 75th percentile. The whiskers show the 10th percentile and the 90th percentile. Notice that TLR 1 had the highest median value. TLR 7 was present in all HNLs. TLR9 shows a significant difference between the polyclonal and the oligoclonal cases.
PCR for 16S rDNA of Bacterial Origin
The test was done on all 24 cases of HNL, with genomic DNA from 10 different bacterial species as positive controls, and 10 reactive lymph nodes as negative controls.
We used two pairs of primers to target conserved regions of all bacterial 16S rDNAs. The first pair was 5′-(6FAM)-T(A/T)A-N(G/A)C-ATG-CAA-GTC-G-3′ (49–64) and 5′-TTAC(G/T)CACCCGTN(T/C)GCC-3′ (120–104).40 The second pair was 5′-GGTTAAGTCCCGCAACGAGCGCAAC-3′ (1088-1112) and 5′-(6FAM)-GTAAGGGCCATGA(T/G)GACTTGACGTC-3′ (1214–1190).41 The numbers in parentheses indicate the corresponding nucleotide positions in Escherichia coli 16S rDNA.
Specifically, a 20 μl PCR mixture included 0.1 μg of DNA extracted from formalin-fixed, paraffin-embedded lymph nodes of HNL and 0.3 μM of each primer. The reaction mixture was subjected to 30 cycles of PCR. Each cycle consisted of denaturation at 94°C for 45 s, annealing at 56°C for 45 s, and extension at 72°C for 45 s. The sizes of the PCR products were 72 for the first pair, and 127 for the second pair. A typical result of polyacrylamide gel electrophoresis was shown in Figure 4.
Polyacrylamide gel electrophoresis of PCR products of bacterial 16S rDNA. Two pairs of primers were used to amplify the conserved regions. In all, 10 bacteria were used as positive controls, and 10 reactive lymph nodes as negative controls. Although the tests were done on all 24 HNLs, only parts of the data are shown here. The leftmost lane is the 20-bp marker. Lanes 1 and 2: 2 bacterial samples as positive controls. Lanes 3 and 4: 2 reactive lymph nodes as negative controls. Lanes 5–9: five cases of HNL. Note that the 1st pair of primers (upper panel) appeared to have a higher background, so that the two negative controls were weakly positive. Two HNLs were positive, but not as strongly as the positive controls. The 2nd pair (lower panel) gave a very weak background. The negative controls and the HNLs were all negative. With these two pairs of primers, only two of the 24 HNLs had a positive PCR product, making common bacteria an unlikely etiology.
Immunohistochemistry
Immunoperoxidase stains for CD123 (a marker for pDCs),42 CD14 (a myeloid marker),43 CD83 (a marker of mature DCs),44, 45 TLR9, and CD69 (a marker of activated T cells),46 were done on formalin-fixed, paraffin-embedded tissue sections of all 24 cases. The antibodies against CD14 (NCL-CD14-223), CD83 (1H4B), and CD69 (NCL-CD69) were from Novocastra (Newcastle upon Tyne, UK), the CD123 antibody (9F5) was from BD Pharmingen (San Diego, CA, USA), and the TLR9 antibody was from Asian HepatoGene (Kaoshiung, Taiwan). The staining was done under conditions recommended by the vendor, with tyramine amplification for CD123. Illustrative examples are shown in Figure 5.
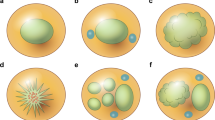
Immunohistochemistry showed rare mature CD83+ DCs (a) CD123+ pDCS rimming the characteristic subcortical necrosis, (b) CD14+ monocytes or mDCs in a subcapsular distribution, (c) rare mature CD83+DCs in the interstitium, (d) CD123+ pDCs in a confluent pattern next to a focus of necrosis, (e) TLR9+ pDCs next to a focus of necrosis in a case with OTCs, and (f) rare CD69+T cells in the interstitium. Immunoperoxidase stains on formalin-fixed, paraffin-embedded tissue, × 40 (a–c), × 200 (d–f).
Results
Frequent Oligoclonal T-Cell Populations in HNL
Oligoclonal T-cell expansion in response to specific antigens has been well documented in infectious and autoimmune diseases.6 Consistent with our previous data, we found frequent oligoclonal T-cell populations in the present study (12/24 or 50%, Table 3). This suggested a restricted immune response in HNL.5
Universal Presence of TLR7 Confirms the Presence of pDCs
HNL is characterized by the presence of plasmacytoid dendritic cells (pDCs). Unlike myeloid dendritic cells (mDCs) that express a broad spectrum of TLRs, pDCs express mainly TLR7 and TLR9. It is therefore of interest to see if the patterns of TLR transcripts in HNL reflect the universal presence of pDCs.
In Table 3, the 24 HNLs were divided into an oligoclonal group and a polyclonal group. Regardless of T-cell clonality, there was indeed a universal presence of TLR7, which is a TLR associated with pDCs for the recognition of single-stranded RNA. This confirmed the presence of pDCs, which is a characteristic finding in HNL.
Oligoclonal T Cells Correlate with TLR9 Transcripts but not with Clinical Manifestation
Classically, the major functions of DCs are to capture antigens, process them through the MHC II pathway, and present them to stimulate CD4+ helper T cells.10, 11 Stimulation of CD8+ cytotoxic T cells through the MHC I pathway is possible, although the mechanism underlying this ‘cross presentation’ is still incompletely understood.29, 30, 31 Because most T lymphocytes in HNL are CD8+ cytotoxic T cells, it is interesting to determine if there is a correlation between cytotoxic T-cell proliferation and the TLRs used by DCs.
In Table 3, we found a correlation between oligoclonal T cells and the presence of TLR9 (at P=0.02), which was also the only TLR found to be significantly correlated with T-cell clonality in a stepwise regression analysis based on a logistic model (P=0.02). Because TLR9 is usually expressed by pDCs, this association strongly suggested expansion of oligoclonal T cells by TLR9+ pDCs, probably in response to unmethylated CpG-rich DNA of bacterial or viral origin.
Figure 3 shows the distribution of TLRs in the oligoclonal vs polyclonal group in a box–whisker plot, which represents the 10, 25, 50, 75, and 90 percentiles of the TLRs. We found that the box for the oligoclonal group was significantly higher than the box for the polyclonal group only for TLR9 (P=0.002, Mann–Whitney test). TLR9, a pDC-associated TLR, was thus the only TLR whose distribution was significantly different between the oligoclonal and the polyclonal groups. This confirmed the conclusion of Table 3.
Of note, there is no association between T-cell clonality and clinical manifestations in Table 3.
Abundant TLR1 Transcripts of Unknown Significance
In Figure 3, we noticed that TLR1 had the highest median value among all TLRs. The abundant TLR1 transcripts in HNL might be due to its broad distribution in dendritic cells and lymphocytes, but further studies are required for a proper interpretation.
Negative PCRs for Bacterial 16S rDNA Favor a Viral Etiology for HNL
Since TLR9 is the receptor for unmethylated CpG-rich DNA of bacterial or viral origin, we used PCRs to look for 16S rDNA of bacterial origin. To increase the sensitivity for detecting bacterial 16S rDNA, we used two pairs of primers. However, only two of the 24 HNLs had a positive PCR product (Figure 4), making common bacteria an unlikely etiology. Alternatively, a virus might be the causative agent of HNL.
Immunohistochemistry (IHC) Confirmed the Presence of CD123+ pDCs that Lack the Maturation Marker CD83
We have used IHC to define the pathology of HNL. IHC for CD123, a marker for pDCs,42 showed the presence of CD123+ pDCs surrounding the subcortical necrosis in every case of HNL (Figure 5a). IHC for CD14 was used as a surrogate marker for monocytes or mDCs,43 and showed their presence in a subcapsular pattern in most cases (Figure 5b). In contrast to the strong staining for CD123 and CD14, IHC for CD83,44, 45 a marker for mature DCs, was rarely positive (Figure 5c). The presence of immature DCs, that had only weak stimulatory activity, might account for the relative benign course of HNL.
CD123+ pDCs may also be found in a confluent pattern (Figure 5d). A significant number of these CD123+ pDCs were also TLR9+ in the cases with OTCs (Figure 5e), but only rare T cells were positive for the activation marker CD69 (Figure 5f). Again, this lack of the activation marker might explain the benign clinical course of HNL.
Discussion
Kikuchi's lymphadenitis (histiocytic necrotizing lymphadenitis, HNL) is a self-limiting lymphadenopathy of unknown etiology. It is characterized by subcortical patchy necrosis with infiltrating pDCs and often oligoclonal cytotoxic T cells, but no neutrophils.3, 4, 5 Although the diagnosis is usually apparent in an endemic area like Taiwan, the etiology is elusive and data on the interaction between pDCs and T cells are limited, making the diagnosis sometimes difficult in atypical cases with a protracted clinical course.
To resolve these problems, we completed an extensive analysis of the TLR repertoire in the lymph nodes. We confirmed the universal presence of TLR7 pDCs that lacked the maturation marker CD83, and found a strong correlation between oligoclonal T-cell proliferation and the presence of TLR9+ pDCs.
The presence of TLR+ pDC in HNL is not unexpected, as early studies had shown increased infiltration of plasmacytoid monocytes in HNL. Plasmacytoid monocytes are now known as pDCs. They respond to certain types of bacteria and viruses by secreting high levels of IFN-α, -β, and -ω, or by upregulating costimulatory molecules such as CD80, CD83, and CD86.2, 3 The presence of immature CD83− DCs in HNL, that had only weak stimulatory activity, is thus significant and might account for the relatively benign course of HNL.
The correlation between oligoclonal T-cell proliferation and the presence of TLR9+ pDCs is consistent with reports that TLR9 is a potent stimulator and an essential component for cross priming of cytotoxic T cells.29, 30, 31 TLR9+ pDCs are activated by single-stranded oliogodeoxynucleotides containing CpG motifs (CpG ODNs).47 Depending on the specific forms of CpG ODNs, pDCs show distinct immune maturation pathways, one of which leads to limited pDC maturation but enhanced IFN-α/β secretion, resulting in the stimulation of naïve CD8 cells.48 Our finding of TLR9+ immature pDCs in association with oligoclonal T cells is consistent with this scenario.
pDCs express TLR7 and TLR9. TLR7 and TLR9, together with TLR8, form a functional subgroup that recognizes pathogen-associated molecular patterns in the endosomal/lysosomal compartments.49 Bacteria/viruses initially interact with cell-surface receptors. After internalization, the bacterial/viral DNA may be digested and released in an endosomal compartment for interaction with TLR9. However, the activation of endosomal TLR9 does not distinguish a bacterial from a viral cause. The negative PCRs for 16S bacterial rDNA and previous reports of repeated negative bacterial cultures point to the possibility of a viral etiology for HNL. Several types of viruses, such as human herpesviruses types 4, 6, and 8 (EBV, HHV-6, KSHV/HHV 8), have been postulated in some cases of HNL, but a definitive viral agent has not been identified.50
HNL resembles systemic lupus erythematosus (SLE) in several aspects, despite the very different clinical courses. They have similar morphologies and frequent oligoclonal T cells,51 and are both associated with pDCs and mDCs.52 On the other hand, autoreactive CD4+ T cells, B-cell hyperactivity, vasculitis, and presence of neutrophils are characteristic of SLE, but are rarely found in HNL. Increased levels of hypomethylated CpG-containing DNA in sera from patients with SLE may contribute to the initiation and/or perpetuation of the disease.52 pDCs from SLE patients respond abnormally to CpG ODN stimulation, which may contribute to the cytokine imbalance observed in this disease.52 Recently, it was shown that the TLR 9 pathway is activated in SLE patients, and the activation could induce disease progression in a mouse model.53, 54 A systematic study on the TLR profile in SLE, with the same approach used in this paper, might shed light on its relation to HNL.
The same approach can be applied to other reactive lymphadenopathies. Data are already available on the association of specific TLRs with infectious agents, such as TLR2 in mycobacterial and herpes viral infections.55, 56 However, comprehensive studies on the TLR profiles as diagnostic or prognostic indicators in these diseases are still lacking. Of particular interest might be the TLR profiles in other enigmatic diseases with involvement of DCs, such as sarcoidosis, Castleman’s disease, and Kimura’s disease. We have, indeed, found the presence of both pDCs and TLR9 in Kimura’s disease (data not shown).
Besides the RT-PCR approach used in this report, the IHC of TLRs and DCs is of equal importance. Such data are available for chronic gastritis and chorioamnionitis,57, 58 but not for reactive lymphadenopathies. We have performed IHC for CD123, a marker for pDCs, on a variety of reactive lymphadenopathies. Extensive infiltrates of CD123+ pDCs are rarely found outside Kikuchi’s disease (data not shown). The presence of pDCs might be used to separate Kikuchi’s disease from T-cell lymphoma.5
In conclusion, by analyzing TLR repertoires, we found the presence of TLR7 pDCs in HNL. The lack of CD83, a marker for mature DCs, implies weak stimulation of T cells. However, the presence of TLR9+ pDCs correlates with oligoclonal T-cell proliferation. These findings echo the recent finding of cross-priming of cytotoxic T cells by pDCs and confirm the primary role of pDCs in the pathogenesis of HNL. Since the natural ligand for TLR9 is hypomethylated CpG-rich DNA of microbial origin, the absence of bacterial 16S rDNA indirectly suggests a viral etiology for HNL.
References
Kikuchi M . Lymphadenitis showing focal reticulum cell hyperplasia with nuclear debris and phagocytes. Acta Hematol Jpn 1972;35:379–380.
Fujimoto Y, Kojima Y, Yamaguchi K . Cervical subacute necrotizing lymphadenitis. Naika 1972;30:920–927.
Facchetti F, de Wolf-Peeters C, van den Oord JJ, et al. Plasmacytoid monocytes (so-called plasmacytoid T-cells) in Kikuchi's lymphadenitis. Am J Clin Pathol 1989;92:42–50.
Pileri SA, Facchetti F, Ascani S, et al. Myeloperoxidase expression by histiocytes in Kikuchi's and Kikuchi-like lymphadenopathy. Am J Pathol 2001;159:915–924.
Lin CW, Chang CL, Li CC, et al. Spontaneous regression of Kikuchi’s lymphadenopathy with oligoclonal T-cell populations favors a benign immune response over a T-cell lymphoma. Am J Clin Pathol 2002;117:627–635.
Maini MK, Casorati G, Dellabona P, et al. T-cell clonality in immune responses. Immunol Today 1999;20:262–266.
Shortman K, Liu YJ . Mouse and human dendritic cell subtypes. Nat Rev Immunol 2002;2:151–161.
Banchereau J, Briere F, Caux C, et al. Immunobiology of dendritic cells. Ann Rev Immunol 2000;18:767–811.
Janeway CA, Medzhitov R . Innate immune recognition. Ann Rev Immunol 2002;20:197–216.
Lanzavecchia A, Sallusto F . Regulation of T cell immunity by dendritic cells. Cell 2001;106:263–266.
Guermonprez P, Valladeau J, Zitvogel L, et al. Antigen presentation and T cell stimulation by dendritic cells. Ann Rev Immunol 2002;20:621–667.
Lennert K, Remmele W . Karyometrische Untersuchungen an Lymphknotenzellen des Menschen: I. Mitt. Germinoblasten, Lymphoblasten und Lymphozyten. Acta Haematol (Basel) 1958;19:99–113.
Facchetti F, de Wolf-Peeters C, Mason DY, et al. Plasmacytoid T cells, immunohistochemical evidence for their monocyte/macrophage origin. Am J Pathol 1988;133:15–21.
Blom B, Ligthart SJ, Schotte R, et al. Developmental origin of Pre-DC2. Hum Immunol 2002;63:1072–1080.
Corcoran L, Ferrero I, Vremec D, et al. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J Immunol 2003;170:4926–4932.
Banchereau J, Pulendran B, Steinman R, et al. Will the making of plasmacytoid dendritic cells in vitro help unravel their mysteries? J Exp Med 2000;192:F39–F44.
Takeda K, Kaisho T, Akira S . Toll-like receptors. Annu Rev Immunol 2003;21:335–376.
Ozinsky A, Underhill DM, Fontenot JD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc Natl Acad Sci USA 2000;97:13766–13771.
Hornung V, Rothenfusser S, Britsch S, et al. Quantitative expression of Toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol 2002;168:4531–4537.
Kadowaki N, Ho S, Antonenko S, et al. Subsets of human dendritic cell precursors express different Toll-like receptors and respond to different microbial antigens. J Exp Med 2001;194:863–869.
Zarember KA, Godowski PJ . Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol 2002;168:554–561.
Jarrossay D, Napolitani G, Colonna M, et al. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur J Immunol 2001;31:3388–3393.
Barton GM, Medzhitov R . Toll-like receptors and their ligands. Curr Top Microbiol Immunol 2002;270:81–92.
Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004;303:1526–1529.
Diebold SS, Kaisho T, Hemmi H, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004;303:1529–1531.
Bottcher T, von Mering M, Ebert S, et al. Differential regulation of Toll-like receptor mRNAs in experimental murine central nervous system infections. Neurosci Lett 2003;344:17–20.
Qureshi ST, Medzhitov R . Toll-like receptors and their role in experimental models of microbial infection. Genes Immun 2003;4:87–94.
Barton GM, Medzhitov R . Control of adaptive immune responses by Toll-like receptors. Curr Opin Immunol 2002;14:380–383.
Heit A, Maurer T, Hochrein H, et al. Toll-like receptor 9 expression is not required for CpG DNA-aided cross-presentation of DNA-conjugated antigens but essential for cross-priming of CD8 T cells. J Immunol 2003; 170:2802–2805.
Datta SK, Redecke V, Prilliman KR, et al. A subset of Toll-like receptor ligands induces cross-presentation by bone marrow-derived dendritic cells. J Immunol 2003;17:4102–4210.
Schwarz K, Storni T, Manolova V, et al. Role of Toll-like receptors in costimulating cytotoxic T cell responses. Eur J Immunol 2003;33:1465–1470.
Williams DL, Ha T, Li C, et al. Toll-like receptor 2 and 4 during the early phases of polymicrobial sepsis correlates with mortality. Crit Care Med 2003;31: 1808–1818.
Wiemann S, Weil B, Wellenreuther R, et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res 2001;11:422–435.
Rock FL, Hardiman G, Timans JC, et al. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA 1998;95:588–593.
Medzhitov R, Preston-Hurlburt P, Janeway Jr CA . A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997; 388:394–397.
Chaudhary PM, Ferguson C, Nguyen V, et al. Cloning and characterization of two Toll/interleukin-1 receptor-like genes TIL3 and TIL4: evidence for a multi-gene receptor family in humans. Blood 1998;91:4020–4027.
Takeuchi O, Kawai T, Sanjo H, et al. TLR6: A novel member of an expanding Toll-like receptor family. Gene 1999;231:59–65.
Du X, Poltorak A, Wei Y, et al. Three mammalian Toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw 2000;11:362–371.
Chuang TH, Ulevitch RJ . Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta 2001;1518:157–161.
Bertilsson S, Cavanaugh CM, Polz MF . Sequencing-independent method to generate oligonucleotide probes targeting a variable region in bacterial 16S rRNA by PCR with detachable primers. Appl Environ Microbiol 2002;68:6077–6086.
Greisen K, Loeffelholz M, Purohit A . PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol 1994;32:335–351.
Summers KL, Hock BD, McKenzie JL . Phenotypic characterization of five dendritic cell subsets in human tonsils. Am J Pathol 2001;159:285–295.
Setoguchi M, Nasu N, Yoshida S, et al. Mouse and human CD14 (myeloid cell-specific leucine-rich glycoprotein) primary structure deduced from cDNA clones. Biochim Biophys Acta 1989;1008:213–222.
Berchtold S, Muhl-Zurbes P, Heufler C, et al. Cloning, recombinant expression and biochemical characterization of the murine CD83 molecule, which is specifically upregulated during dendritic-cell maturation. FEBS Lett 1999;461:211–216.
Scholler N, Hayden-Ledbetter M, Dahlin A, et al. Cutting edge: CD83 regulates the development of cellular immunity. J Immunol 2002;168:2599–2602.
Lopez-Cabrera M, Santis AG, Fernandez-Ruiz E, et al. Molecular cloning, expression, and chromosomal localization of the human earliest lymphocyte activation antigen AIM/CD69, a new member of the C-type animal lectin superfamily of signal-transmitting receptors. J Exp Med 1993;178:537–547.
Krieg AM . CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol 2002;20:709–760.
Verthelyi D, Zeuner RA . Differential signaling by CpG DNA in DCs and B cells: not just TLR9. Trends Immunol 2003;24:519–522.
Heil F, Ahmad-Nejad P, Hemmi H, et al. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur J Immunol 2003;33:2987–2997.
Hudnall SD . Kikuchi-Fujimoto disease. Is Epstein–Barr virus the culprit? Am J Clin Pathol 2000;113:761–764.
Holbrook MR, Tighe PJ, Powell RJ . Restrictions of T cell receptor beta chain repertoire in the peripheral blood of patients with systemic lupus erythematosus. Ann Rheum Dis 1996;55:627–631.
Pascual V, Banchereau J, Palucka AK . The central role of dendritic cells and interferon-alpha in SLE. Curr Opin Rheum 2003;15:548–556.
Boule MW, Brought C, Mackay F, et al. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med 2004;199:1631–1640.
Anders HJ, Vielhauer V, Eis V, et al. Activation of toll-like receptor-9 induces progression of renal disease in MRL-Fas(lpr) mice. FASEB J 2004;18:534–536.
Gehring AJ, Dobos KM, Belisle JT, et al. Mycobacterium tuberculosis LprG (Rv1411c): a novel TLR-2 ligand that inhibits human macrophage class II MHC antigen processing. J Immunol 2004;173:2660–2668.
Morrison A . The Toll of herpes simplex virus infection. Trends Microbiol 2004;12:353–2356.
Schmausser B, Andrulis M, Endrich S, et al. Expression and subcellular distribution of toll-like receptors TLR4, TLR5 and TLR9 on the gastric epithelium in Helicobacter pylori infection. Clin Exp Immunol 2004;36:521–526.
Kumazaki K, Nakayama M, Yanagihara I, et al. Immunohistochemical distribution of Toll-like receptor 4 in term and preterm human placentas from normal and complicated pregnancy including chorioamnionitis. Hum Pathol 2004;35:47–54.
Acknowledgements
Research funding was provided through Grants NTUH 93-S006 and NSC92-2320-B002-137 to CW Lin, and NSC-91-2314-B-002-179 and NHRI-EX91-704SL to SM Hsu, from the National Taiwan University Hospital (NTUH), the National Science Council (NSC), and the National Health Research Institute (NHRI), Taiwan. We thank Mr CC Lu for the artwork, the National Taiwan University Center for Genomic Medicine for technical support, and the Center for Biostatistics at NTUH for data analysis. The authors have no duality of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, CW., Liu, TY., Lin, CJ. et al. Oligoclonal T cells in histiocytic necrotizing lymphadenopathy are associated with TLR9+ plasmacytoid dendritic cells. Lab Invest 85, 267–275 (2005). https://doi.org/10.1038/labinvest.3700201
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700201