Abstract
The hepatocyte growth factor (HGF) has been crucially implicated in the development of proliferative retinal diseases; however, it is unclear whether retinal glial cells express or respond to HGF. Therefore, we examined the expression of HGF and of the receptor for HGF, c-Met, by immunohistochemical costaining with glial fibrillary acidic protein (GFAP) in epiretinal membranes of patients with proliferative vitreoretinopathy (PVR) and proliferative diabetic retinopathy (PDR), respectively. Furthermore, it was determined whether cells of the human retinal glial cell line, MIO-M1, secrete HGF protein, and whether HGF stimulates proliferation and chemotaxis, and secretion of the vascular endothelial growth factor (VEGF). Neuroretinas of patients with PVR express elevated mRNA level for HGF in comparison to control retinas. In epiretinal membranes of patients with PVR or PDR, immunoreactivity for HGF and for c-Met, respectively, partially colocalized with immunoreactivity for GFAP. Fetal bovine serum and basic fibroblast growth factor, but not heparin-binding epidermal or platelet-derived growth factors, evoked HGF secretion by cultured retinal glial cells. HGF displayed only a marginal effect on cell proliferation while it stimulated chemotaxis. HGF promoted the secretion of VEGF, via activation of the phosphatidylinositol-3 kinase. It is concluded that glial cells in epiretinal membranes express both HGF protein and c-Met receptors. The results suggest an autocrine/paracrine role of HGF in glial cell responses during proliferative vitreoretinal disorders as well as in retinal neovascularization, by stimulating of VEGF release.
Similar content being viewed by others
Main
Complex interactions of retinal cells with growth factors and cytokines, extracellular matrix proteins and metalloproteinases underlie the development of proliferative retinal diseases.1, 2, 3 Among the growth factors present in significant amounts in fibrocellular membranes and in the vitreous fluid of patients with proliferative vitreoretinopathy (PVR), particularly the hepatocyte growth factor (HGF) and the platelet-derived growth factor (PDGF) have been implicated in the development of proliferative retinopathies.4, 5, 6, 7, 8, 9 In addition to retinal pigment epithelial (RPE) and inflammatory cells, retinal glial cells participate in the formation of fibroproliferative tissue associated with PVR and proliferative diabetic retinopathy (PDR),10, 11, 12 and similar to RPE cells, retinal glial cells are immediately activated during experimental retinal detachment and begin to proliferate within hours of detachment.13, 14 Gliotic Müller cells from PVR retinas display distinct changes in their physiological and morphological features compared to cells from post-mortem donors. They show, for example, cellular hypertrophy and express immunoreactivity for glial fibrillary acidic protein (GFAP),15 and they display altered expression and responsiveness of cell surface receptors.
HGF has been crucially implicated in the development of proliferative retinopathies as it induces scattering and migration of RPE cells, and also displays a slight mitogenic effect in these cells.6, 16, 17 However, it is unknown until today whether, besides RPE cells, retinal glial cells express or respond to HGF. In a rat model of transient ischemia, HGF has been described to be expressed in the inner retina within 24 h of reperfusion, probably by Müller cells,18 suggesting a role of this factor in gliotic responses upon ischemic/hypoxic injury. We carried out immunohistochemical staining to localize HGF protein and the c-Met receptor for HGF in glial cells of surgically excised epiretinal membranes of patients with PVR and PDR, and determined whether different growth factors and cytokines as well as serum may evoke the release of HGF from retinal glial cells. In order to reveal a physiological significance of c-Met receptor expression, the HGF-evoked secretion of the vascular endothelial growth factor (VEGF) by retinal glial cells was investigated.
Materials and methods
Materials
The human recombinant growth factors and cytokines heparin-binding epidermal growth factor-like growth factor (HB-EGF), HGF, basic fibroblast growth factor (bFGF), and platelet-derived growth factor (PDGF-BB) were purchased from R&D Systems (Minneapolis, MN, USA). LY294002 was obtained from Calbiochem (Bad Soden, Germany), and UO126 and SB203580 were from Tocris (Ellisville, MO, USA). All other substances were obtained from Sigma (Deisenhofen, Germany). The following antibodies were used: mouse anti-human HGF (1:100; Sigma), rabbit anti-human β-chain of c-Met (1:500; Acris, Herford, Germany), rabbit anti-bovine GFAP (1:1000; Dako A/S, Glostrup, Denmark), Cy2- or Cy3-conjugated goat anti-mouse IgG (1:400; Dianova), and Cy2- or Cy3-coupled goat anti-rabbit IgG (1:400; Dianova).
Ocular Tissues
All tissue was used in accordance with applicable laws and with the Declaration of Helsinki for research involving human tissue, and was approved by the ethics committee of the Leipzig University Medical School. For oligonucleotide microarray analysis and reverse transcriptase-polymerase chain reaction (RT-PCR), respectively, the total RNA of 10 post-mortem retinas of human subjects with no reported history of eye disease (four women, six men; age 64.4±18.2 years; range, 17–86 years) were used (post-mortem time up to 24 h). Additionally, the total mRNA from retinal fragments of seven patients undergoing 360-grade retinectomies for PVR (grades CP4, CA4 and above) was analyzed (five women, two men; age 72.4±2.9 years; range, 68–76 years). In all, 12 surgically excised epiretinal membranes (PVR or PDR), obtained from consenting patients during vitrectomy surgery (seven women, five men; age 67.7±9.8 years; range, 44–87 years), were used for immunohistochemical staining.
Total RNA Preparation
Immediately after surgery, PVR retinas were removed from the vitrectomy waste, and precipitated by centrifugation at 4°C, washed twice with prechilled phosphate-buffered saline to remove blood cells, and used for total RNA preparation. The preparation was carried out using TRIZOL (Gibco BRL, Paisley, UK); 1 ml TRIZOL was added to the tissue material and homogenized using an Ultraturrax (IKA, Staufen, Germany). The homogenized samples were incubated at room temperature for 5 min, and after addition of 0.2 ml chloroform, the samples were mixed vigorously and incubated for 2–3 min at room temperature (RT). Thereafter, RNA was additionally purified by using the RNeasy Mini Kit (Qiagen, Hilden, Germany). After centrifugation (12 000 × g, 15 min at 4°C), the RNA-containing phase was transferred into a fresh tube, and total RNA was precipitated by mixing with 0.5 ml isopropyl alcohol followed by an incubation for 10 min at RT. After centrifugation, the RNA pellets were washed twice with 75% ethanol, air dried and, subsequently, dissolved in RNase-free water. The samples were analyzed by electrophoresis to assure that the ribosomal RNA was intact. The purity and quantity of RNA was estimated by measuring the optical density (OD) ratio OD260/OD280 by using the GeneQuantpro device (Pharmacia, Uppsala, Sweden) which was between 1.9 and 2.1 for all samples, indicating that the RNA samples were of sufficient quality.
Oligonucleotide Microarray Analysis
The expression of different types of mRNA in the total mRNA preparations was analyzed by using the Human Genome U95Av2 array of the Affymetrix Genechip System (Affymetrix, Santa Clara, CA, USA), according to the manufacturer's instructions. In this microarray, each gene analyzed was represented by 16–20 different oligonucleotide probe pairs where each probe pair consisted of a match and a mismatch oligonucleotide. The mismatch probes, which served as controls for the determination of background and of nonspecific hybridization signals, included a single base substitution which inhibited the hybridization with the mRNA of the target gene. The mRNA expression levels were evaluated using the Affymetrix Microarray Suite 5.0 software that calculated three detection levels (present, marginal, absent) and the size of the messages, by considering both the intensities of the signals that were emitted from the probe sets and the number of probe pairs in which the perfect match was specific. A certain species of mRNA was defined as being expressed when the difference between the perfect match signal and the mismatch signal was significant (P<0.05). A comparison analysis was carried out, which evaluated the relative change in abundance for each transcript between a baseline (normal retina) and a experimental sample (each PVR retina). The integrity of the used cRNA samples was tested by checking the 5′-to-3′ratios of the housekeeping genes.
RT-PCR
cDNA was synthesized from 1 μg total RNA using the First-Strand cDNA Synthesis Kit for RT-PCR (Roche, Mannheim, Germany). For PCR, the Taq PCR Master Mix Kit (Qiagen, Hilden, Germany) was used. Amounts of 1 μl of the first-strand mixture and 1 μM of each gene-specific sense and antisense primer were used for the amplification reaction in a final volume of 50 μl. The following primer pairs were used: HGF (Accession No. X16323), sense 5′-AGGAGAAGGCTACAGGGGCAC-3′, antisense 5′-TTTTTGCCATTCCCACGA-TAA-3′, producing a 267 bp amplicon; c-Met (Accession No. NM_000245), sense 5′-TGAAGTCATAGGAAGAGGGCA-3′, antisense 5′-GGCAAGACCAAAATCAGCAAC-3′, producing a 436 bp amplicon; β-actin (Accession No. M10277), sense 5′-ATGGCCACGGCTGCTTCCAGC-3′, antisense 5′-CATGGTGGTGCCGCCAGACAG-3′, producing a 237 bp amplicon; GAPDH (Accession No. M33197), sense 5′-GCAGGGGGGAGCCAAAAGGGT-3′, antisense 5′-TGGGTGGCAGTGATGGCATGG-3′, producing a 219 bp amplicon. Thermocycling was carried out by using the PTC-200 Thermal Cycler (MJ Research, Watertown, MA, USA), according to the following protocol: denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and polymerization at 72°C for 2 min. A final extension step was made at 72°C for 10 min. The amplified products were separated on a 2% agarose gel containing 10 ng/ml ethidium bromide, and were visualized using the ultraviolet transilluminator of the Fluor-S-Imager (BioRad, Munich, Germany).
Quantitative Real-Time PCR
The total mRNA preparations from control and PVR retinas were analyzed in triplicate and duplicate, respectively. The contaminating genomic DNA was eliminated by using 1 U of DNase I (Roche, Mannheim, Germany) per μg of total RNA. First-strand cDNA synthesis was performed with 1 μg of total RNA by using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA, USA) in a final volume of 20 μl. The following primer pairs were used: HGF (Accession No. X16323): sense 5′-GGCTGGGGCTACACTGGATTG-3′, antisense 5′-CCACCATAATCCCCCTCACAT-3′, producing a 179 bp amplicon; and GAPDH (see above). Real-time PCR was performed with the Light Cycler (Roche Diagnostics, Indianapolis, IN, USA). The PCR solution contained 1 μl cDNA, specific primer set (0.3 μM each), 3 mM MgCl2, and 2 μl of SYBR Green Master Mix (Light Cycler DNA Master SYBR Green I Kit, Roche) in a final volume of 20 μl. The PCR parameters were initial denaturation, one cycle at 95°C for 5 s; amplification and quantification, 40 cycles at 95°C for 4 s, 58°C for 5 s, and 72°C for 7 s; melting curve, 1 cycle at 65°C with the temperature gradually increased up to 95°C (temperature transition, 0.2°C/s). Relative quantification of real-time PCR results was performed by using the mathematical model of Pfaffl.19 The changes in HGF gene expression were calculated by using the average values of three independent experiments; the HGF mRNA expression was normalized to the levels of GAPDH mRNA. The degree of change in HGF mRNA was calculated and compared with the control retina; results are expressed as means±s.e.m.
Cell Culture
The culture experiments were carried out using the spontaneously immortalized human Müller cell line, MIO-M1.20 The cells were cultured in tissue culture flasks (Greiner, Nürtingen, Germany) in Dulbecco's modified Eagle's medium (Invitrogen, Paisley, UK) containing 10% fetal bovine serum, glutamax II, and gentamycin in 19.5% O2/5% CO2 at 37°C. To carry out the proliferation experiments, the cells were seeded at 3 × 103 cells per well in 96-well-flat-bottom microtiter plates (Greiner) that contained medium supplemented with 10% serum, and were allowed to attach for 48 h. Thereafter, the cells were growth-arrested in medium without serum for 16 h, and subsequently, the test substances were added to the culture medium for another 24 h.
DNA Synthesis Rate
The DNA synthesis rate was assayed by measuring the bromodeoxyuridine (BrdU) incorporation using the Cell Proliferation ELISA BrdU Kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. BrdU (10 μM) was added to the culture medium 5 h before fixation of the cells with 4% paraformaldehyde.
Chemotaxis
Measurements of chemotaxis were performed by using a modified Boyden chamber assay. Suspensions of MIO-M1 cells (100 μl; 5 × 105 cells/ml serum-free medium) were seeded onto polyethylene terephthalate filters (pore size 8 μM; Becton Dickinson, Heidelberg, Germany) coated with fibronectin (50 μg/ml) and gelatin (0.5 mg/ml). Within 4 h after seeding, the cells attached to the filter and formed a semiconfluent monolayer. The medium was then changed into medium without additives in the upper well and medium containing HGF at different concentrations in the lower well. After incubation for 16 h, the inserts were washed with buffered saline, fixed with Karnofsky's reagent, and stained with hematoxylin. Nonmigrated cells were removed from the filters by gentle scrubbing with a cotton swab. The migrated cells were counted, and the results were expressed relative to cell migration without HGF. Experiments were repeated at least three times.
ELISA
MIO-M1 cells were cultured at 8 × 104 cells per well in six well plates (1.5 ml culture medium per well). After ∼80% confluency was achieved, the cells were cultured in serum-free medium for 16 h. Subsequently, the culture medium was changed, and the cells were stimulated by growth factors and cytokines as well as serum at the doses indicated, in the absence and presence of blocking substances. The supernatants were collected at 24 h, and levels of VEGF-A165 or HGF in the supernatants (200 and 50 μl, respectively) were determined by ELISA (R&D Systems). The cell viability was measured by trypan blue dye exclusion; no significant differences to control were found (not shown).
Immunohistochemistry
Epiretinal membranes were fixed in acetone, and stored at −80°C. The membranes were immunostained as free floating wholemounts. After incubation in 10% normal goat or donkey serum plus 0.3% Triton X-100 in saline for 1 h, the whole mounts were incubated in primary antibody overnight at 4°C. After washing in 1% bovine serum albumin in saline, the secondary antibodies were applied for 4 h at room temperature. The labeling was visualized by means of a confocal laser scanning microscope LSM 510 (Zeiss, Oberkochen, Germany). Control stainings with omitting the anti-HGF or anti-c-Met antibodies revealed no immunoreactivity (not shown).
Statistics
BrdU incorporation and migration rates are expressed as percent of untreated control (100%). Data are expressed as means±s.e.m.; statistical significance (Student's t-test) was accepted at P<0.05.
Results
HGF mRNA Expression in Retinas
In order to examine whether the mRNA of HGF is expressed in human retinas, the RT-PCR analysis was carried out by using the total mRNA extracted from the retinas of four patients with PVR. As shown in the inset of Figure 1a, all retinas investigated expressed the mRNA for HGF. To obtain more quantitative data on the mRNA expression level, a genechip microarray analysis was carried out on the total mRNA of seven post-mortem control retinas of individuals without reported eye disease and two retinal fragments ectomized from patients with PVR. In the post-mortem control retinas, the expression signals of HGF mRNA was small; similarly, the signals for the mRNAs of various other factors (eg, tumor necrosis factor-α and transforming growth factor-β) were below the significance level of the method used. When compared to post-mortem control retinas, the mRNA levels of transforming growth factor-β and HGF among other factors were found to be higher in the retinectomy material from patients with PVR (Figure 1a). The mRNA of HGF was approximately six-fold stronger expressed in PVR retinas than in control retinas. To validate the cDNA microarray results, the total RNA was prepared from post-mortem control and PVR retinas, and the expression levels of HGF mRNA were examined by real-time RT-PCR. By using this method, a three-fold higher expression level of HGF mRNA was observed in PVR retinas compared to control (Figure 1b). The increased expression of mRNA for HGF in PVR retinas compared to post-mortem control retinas suggests that the factor may be involved in alterations of retinal physiology during PVR.
The level of HGF mRNA is increased in retinas of patients with PVR compared to post-mortem control retinas. (a) Relative mRNA levels of different growth factors and cytokines in retinectomy material of patients with PVR as compared to control retinas (100%). The mRNA levels were determined by a genechip microarray for the following factors: aFGF, acidic fibroblast growth factor; bFGF, basic fibroblast growth factor; HGF, hepatocyte growth factor; IGF-1, insulin-like growth factor-1; PDGF-A, platelet-derived growth factor (A chain); TGF-β, transforming growth factor-β; TNF-α, tumor necrosis factor-α. The mRNA of 360-grade ectomized retinas from two patients with PVR was analyzed; the control value of mRNA expression was obtained from the total mRNA of seven post-mortem retinas. (Inset) In four different PVR retinas, the presence of mRNA for HGF could be demonstrated by RT-PCR. (b) Real-time RT-PCR revealed higher expression levels of HGF mRNA in PVR retinas compared to control retinas.
HGF Immunoreactivity in Epiretinal Membranes
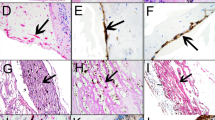
In order to evaluate whether HGF immunoreactivity is expressed by glial cells in epiretinal membranes, whole mounts of surgically excised membranes were costained for HGF and the glial cell marker GFAP. We investigated three fibrocellular and three fibrovascular epiretinal membranes of patients with PVR and PDR, respectively. All membranes investigated showed positive staining for both HGF and GFAP. In PVR membranes, there were large regions which were costained with both antibodies and, therefore, displayed a yellow fluorescence in the overlay (right side of Figure 2a, b). Additionally, there were also GFAP-negative regions that were stained for HGF (Figure 2b). In the PDR membranes investigated, there were often large GFAP-expressing structures which were strongly labelled with the anti-HGF antibody (Figure 2c and left side of Figure 2d), as well as regions where HGF and GFAP imunoreactivities showed no coexpression (right side of Figure 2d). It is concluded that a subpopulation of glial cells in epiretinal membranes, or distinct subcellular regions of glial cells, express HGF protein.
HGF, c-Met and GFAP immunoreactivity in surgically excised fibroproliferative membranes from patients with PVR (a, b, e) and PDR (c–f), respectively. The epiretinal membranes were labeled with antibodies against HGF or c-Met, respectively, and GFAP. In the overlay, colabeling of HGF or c-Met and GFAP immunoreactivity resulted in yellow staining. The membranes were derived from six different patients. Similar staining patterns were observed in 12 different membranes. Scale bars, 20 μM.
c-Met Immunoreactivity in Epiretinal Membranes
c-Met immunoreactivity was found in all investigated epiretinal membranes of patients with PVR (Figure 2e) and PDR (Figure 2f). The immunoreactivity was partially colocalized with GFAP; however, there were also GFAP-expressing regions in the membranes which were not stained for c-Met. Figure 2f shows GFAP labeling of structures around and beside a blood vessel; apparently, perivascular glial cells expressed c-Met immunoreactivity. The results suggest that subpopulations of glial cells or subcellular regions of glial cell bodies in epiretinal membranes express the receptor for HGF, c-Met.
c-Met mRNA in Retinas and Müller Cells
In order to support the assumption that retinal glial cells may express c-Met, RT-PCR studies were carried out in cells of the human Müller cell line, MIO-M1.20 As positive control, the presence of c-Met mRNA in cultured human RPE cells was confirmed (Figure 3). At control conditions, Müller cells expressed c-Met mRNA at a very low level; however, stimulation with basic fibroblast growth factor (bFGF) caused a marked increase in mRNA expression (Figure 3). PVR retinas displayed a higher level of c-Met mRNA expression as compared to post-mortem control retinas (Figure 3). The results suggest that Müller glial cells may express c-Met, and that the expression is upregulated after cytokine stimulation.
Detection of c-Met mRNA in human retinas and in cells of the human Müller cell line, MIO-M1, by RT-PCR. The levels of c-Met mRNA are higher in MIO-M1 cells stimulated with bFGF (10 ng/ml) for 24 h, as compared to untreated control, and in PVR retinas compared to post-mortem control retinas. As positive control, the presence of c-Met mRNA in cultured human retinal pigment epithelial cells is shown. The negative control was done by omitting of cDNA.
Release of HGF by Müller Cells
The partial colocalization of HGF protein and GFAP in fibroproliferative tissues may suggest that glial cells, besides binding of HGF, may produce and secrete this growth factor. In order to examine this assumption, cultured cells of the human Müller cell line, MIO-M1, were stimulated with different growth factors and serum, respectively, and the HGF content in the culture supernatants was measured by ELISA. As shown in Figure 4, addition of bFGF to the culture media caused a significant increase of HGF in the supernatants as compared to the unstimulated control (P<0.05). Since bFGF (at 10 ng/ml) increased the HGF content of the supernatants by ∼180% while it stimulated the cell proliferation by ∼50% (Figure 5) we assume that the increased HGF content was not solely caused by enhanced cell number. A similar strong release of HGF into the culture media was observed in the presence of fetal bovine serum (Figure 4). On the other hand, stimulation of the cells with PDGF or with heparin-binding epidermal growth factor-like growth factor (HB-EGF) did not increase the HGF content of the culture media. The results suggest that Müller cells are capable to produce and secrete HGF, in response to certain growth factors and to serum.
Growth factor- and serum-induced release of HGF by cells of the human Müller cell line, MIO-M1. The cells were stimulated for 24 h, and the HGF concentration in the cultured media was measured by ELISA. The growth factors were tested at 10 ng/ml, and serum at 2%. Data are expressed as percentage of untreated control (100%) which was between 100 and 250 pg/ml. Means±s.e.m. of 2 independent experiments carried out in duplicate. •P<0.05 vs untreated control.
Effects of HGF on the proliferation and chemotaxis of cells of the human Müller cell line, MIO-M1. (a) Dose-response relationships of bFGF and HGF, respectively, effects on the proliferation rate. The BrdU incorporation rate was measured after a 24-h incubation with the respective factor. Data are expressed in percent of untreated control (100%). Means±s.e.m. of three independent experiments carried out in triplicate. (b) Concentration-dependent chemotaxis toward HGF. The data represent cell migration rates in treated wells relative to that in wells without HGF. Data expressed as means±s.e.m. in two independent cultures in duplicate experiments. *P<0.05; ***P<0.001 vs untreated control.
Proliferation of Müller Cells
In order to determine whether HGF exerts a proliferation-stimulating effect on cells of the human Müller cell line, MIO-M1, different concentrations of the factor were added to the culture media for 24 h, and the BrdU incorporation rate was estimated. Addition of HGF (up to 10 ng/ml) did not significantly stimulate the rate of DNA synthesis while at a concentration of 100 ng/ml, HGF slightly increased the proliferation rate (P<0.05) (Figure 5a). Similarly, VEGF (100 ng/ml) did not change the cell proliferation (102.6±6.0%, n=2; P>0.05). In contrast, addition of bFGF to the culture medium evoked a dose-dependent increase in the proliferation rate of these cells, with an apparent EC50 of ∼1 ng/ml and a maximal effect at ∼10 ng/ml. The results suggest that HGF is a rather weak mitogen for cultured Müller cells.
Migration of Müller Cells
HGF has been described to be a motogen for RPE cells and brain astrocytes, respectively.16, 17, 21 In order to reveal whether HGF has a chemotactic effect on Müller glial cells, MIO-M1 cells were stimulated with different concentrations of HGF, and the migration rate was measured after 16 h. As shown in Figure 5b, HGF induced a dose-dependent enhancement of Müller cell migration, with maximal response at 10 ng/ml.
HGF-Evoked Release of VEGF
Retinal hypoxia is a consistent feature of proliferative retinopathies,22 and Müller cells release VEGF in response to hypoxic stimuli.23, 24, 25. Since HGF is expressed by glial cells in fibroproliferative epiretinal membranes (Figure 2), we investigated whether HGF may be a factor that causes VEGF release by Müller cells and whether this effect is mediated by activation of mitogen-activated protein kinases (MAPKs) or of the phosphatidylinositol-3 kinase (PI3 K). MIO-M1 cells were cultured for 24 h in the presence of HGF, and the VEGF content in the supernatants was measured by ELISA. VEGF is constitutively released by the cells into the culture medium; addition of different growth factors (Figure 6a) or of HGF (Figure 6b) to the cultured cells induced an increase of the VEGF concentration in the supernatants. HGF at 50 or 100 ng/ml evoked a doubling of the VEGF release (Figure 6b). At a concentration of 100 ng/ml, HGF evoked a higher and significant increase of the VEGF concentration (by ∼100%) compared to its effect on cell proliferation (by ∼15%) (Figure 5a), suggesting that HGF induces VEGF secretion by Müller cells. Blocking the activation of the extracellular signal-regulated kinases 1/2 (ERK1/2) by the MEK inhibitor UO126 did not diminish the HGF effect while blocking the activation of the p38 MAPK by the selective inhibitor SB203358 slightly decreased the HGF effect (Figure 6c). On the other hand, the inhibitor of the PI3 K, LY294002, significantly (P<0.05) suppressed the HGF-evoked release of VEGF, suggesting that the effect was mainly mediated by HGF-evoked activation of the PI3 K pathway.
Cytokine-induced release of VEGF by cells of the human Müller cell line, MIO-M1. (a) Different growth factors caused an increase of the VEGF content of the culture supernatant. The factors were tested at 10 ng/ml. PDGF, platelet-derived growth factor-BB. HB-EGF, heparin-binding epidermal growth factor-like growth factor. bFGF, basic fibroblast growth factor. (b) The hepatocyte growth factor (HGF) dose-dependently increased the VEGF concentration in the cultured media. The cells were stimulated with different concentrations of HGF for 24 h, and the concentration of VEGF-A165 in the cultured media was measured by ELISA. (c) HGF (50 ng/ml) mediated its effect by stimulation of the phosphatidylinositol-3 kinase (PI3 K) pathway. The following substances were tested: dimethylsulfoxide (DMSO, 0.1%); inhibitors of the activation of mitogen-activated protein kinases, UO126 (4 μM), SB203580 (10 μM); inhibitor of the PI3 K, LY294002 (5 μM). Data are expressed as percentage of untreated control (100%), which was between 100 and 250 pg/ml. Means±s.e.m. of two to three independent experiments carried out in duplicate. *P<0.05, **P<0.01, ***P<0.001 vs untreated control; •P<0.05, significant effect of the blocker.
Discussion
Together with RPE and inflammatory cells, retinal glial cells represent a major cellular constituent of epiretinal membranes of PVR and PDR.10, 11, 12 HGF has been suggested to be one of the main factors stimulating the scattering and the migration of RPE cells in proliferative retinopathies.6, 16, 17 However, nothing is known about a possible involvement of HGF in glial cell responses in proliferative vitreoretinal diseases. We demonstrate here the presence of partial colabeling of HGF and GFAP immunoreactivity in epiretinal membranes of patients with PVR or PDR (Figure 2) that may suggest that glial cells secrete or, at least, bind this factor. However, it is conspicuous that not all of the glial cells present in the membranes expressed HGF immunoreactivity, and that GFAP-expressing structures showed partial co-staining with HGF immunoreactivity and were partially devoid of HGF labeling (Figure 2d). It remains to be determined in future investigations whether the HGF-expressing and GFAP-negative structures in the membranes reflect the expression of HGF protein by other cell types (eg, RPE cells), by glial cells that do not express GFAP, or whether they are HGF protein deposits released by cells into the extracellular matrix. The view that, besides RPE cells, glial cells may constitute one of the sources of HGF in the diseased retina is supported by our observation that, after stimulation with bFGF or serum, cells of the Müller cell line secrete HGF into the extracellular space (Figure 4).
Besides HGF protein, glial cells in fibroproliferative membranes express the receptor for HGF, c-Met, as suggested by the partial colabeling of c-Met and GFAP in PVR and PDR membranes (Figure 2). The level of c-Met mRNA expression in PVR retinas is higher compared to post-mortem control retinas and, in cultured Müller cells, increased after stimulation by bFGF (Figure 3). The increases of cell migration (Figure 5b) and VEGF release by Müller cells after stimulation with HGF (Figure 6b,c) may indicate that these cells express functional c-Met receptors, suggesting that HGF exerts an autocrine/paracrine action on retinal glial cells, as previously suggested for RPE cells.16 In RPE cells, HGF induces an epithelial-to-mesenchymal cell shape alteration and migration of the cells, as well as a modest increase of cellular proliferation.6, 17 In the present study, we found that HGF evoked a slight increase of the Müller cell proliferation only at relatively high concentrations (Figure 5a), while it significantly stimulated the migration of Müller cells (Figure 5b), suggesting that HGF is no (or a weak) mitogen but a motogen for Müller cells. We found that serum and bFGF, but not PDGF or HB-EGF, induced HGF secretion by glial cells (Figure 4). The bFGF-evoked increases in c-Met expression (Figure 3) and HGF release by Müller cells (Figure 4) is also important for understanding of pathological processes that lead to other kinds of hypoxic retinopathies. In an animal model of retinal detachment, which is generally accompanied by hypoxia of the outer retina,26, 27 activation of bFGF-evoked retinal signaling within minutes of detachment has been described.14 One may assume that the bFGF-evoked HGF release may constitute signaling loops between retinal glial and RPE cells, stimulating the migration, morphological alterations, and cytokine release by both types of cells. During vessel leakage, serum entry into the retinal tissue may cause a strong stimulus for HGF secretion by glial cells.
By using GFAP immunostaining of fibroproliferative tissues, it was impossible to discriminate whether Müller glial cells or astrocytes, or both cell types, express HGF and c-Met immunoreactivity. The GFAP-labeled perivascular structures in PDR membranes (Figure 2f) may suggest that, besides activated Müller cells, also retinal astrocytes express c-Met. The MIO-M1 cell line, which was used to investigate physiological responses in the present study, is the only presently available human Müller cell line. It has been shown that MIO-M1 cells retain various different characteristics of primarily isolated Müller cells in culture, for example, electrogenic responses to L-glutamate, and express differentiation markers of Müller cells, among others, cellular retinaldehyde binding protein, glutamine synthetase, and vimentin.20 Despite the absence of high levels of glial fibrillary acidic protein in these cells which may suggest that the cells do not exhibit characteristics of activation in vitro,20 it can not be ruled out that other cell features are different from those observed in cells of the normal healthy retina in situ, for example, a high level of EGF receptor expression.28 However, because of the difficulty in obtaining pure primary Müller cell populations from post-mortem retinal tissue, studies designed to obtain a better understanding of the role of Müller cells in retinal wound healing and proliferative disorders depend upon investigations in Müller cell lines.
In addition to bFGF and other factors, VEGF is one of the major angiogenic factors in the retina involved in the development of PDR.29, 30, 31 Here we show that, beside different cytokines (Figure 6a), HGF increased the release of VEGF by Müller cells (Figure 6c). The HGF-evoked release of VEGF is mediated by activation of PI3 K while activation of ERK1/2 was apparently not involved. In cultured Müller cells, activation of ERK1/2 is crucially implicated in the proliferation-stimulating effect of different growth factors, cytokines, and agonists of G protein-coupled receptors32, 33 while it does not influence the cytokine production and secretion, for example, the release of VEGF evoked by bFGF33 or by HGF (present results). However, inhibition of PI3 K blocks the bFGF-33 and the HGF-evoked secretion of VEGF by Müller cells. Generally, ERKs regulate the gene expression which promotes, among others, the cell proliferation, while activation of PI3 K, via the downstream targets protein kinase B and mTOR, results in stimulation of the protein synthesis at the translational level; the latter pathway is involved in prosurvival signaling cascades.34 It is suggested that HGF is one of the factors that are released into the retina during proliferative vitreoretinal disorders that causes the release of VEGF by glial cells. The glial cell-derived VEGF may be a part of prosurvival signaling in the hypoxic retina; however, it may also contribute to injury-induced retinal degeneration, via inducing neovascularization and leakage of ocular blood vessels, which favor immune cell infiltration and serum entry into the retinal tissue.
The present results suggest that, besides RPE cells, glial cells produce and respond to HGF during proliferative vitreoretinal disorders. A similar involvement of HGF signaling has been described in brain gliosis and glioma tumorigenesis. Both HGF and c-Met have been identified in brain astrocytes, microglial cells, and astrocytic tumors.35, 36, 37 In astrocytic tumor cells, the expression levels of HGF and c-Met correlates with the grade of malignancy,38, 39 and HGF supplementation to cultures enhances the invasiveness of glioma cells while it does not alter their mitogenesis.40 The present results obtained on Müller cells are in agreement with data obtained on cultured brain astrocytes, which revealed that HGF promotes chemokinesis without an influence on mitogenesis.21 However, a dependence of the expression levels of HGF and c-Met in glial cells on the grade of PVR remains to be determined in future investigations.
References
Limb GA, Little BC, Meager A, et al. Cytokines in proliferative vitreoretinopathy. Eye 1991;5:686–693.
Wiedemann P . Growth factors in retinal diseases: proliferative vitreoretinopathy, proliferative diabetic retinopathy, and retinal degeneration. Surv Ophthalmol 1992;36:373–384.
Charteris DG . Proliferative vitreoretinopathy: pathobiology, surgical management, and adjunctive treatment. Br J Ophthalmol 1995;79:953–960.
Cassidy L, Barry P, Shaw C, et al. Platelet derived growth factor and fibroblast growth factor basic levels in the vitreous of patients with vitreoretinal disorders. Br J Ophthalmol 1998;82:181–185.
Andrews A, Balciunaite E, Leong FL, et al. Platelet-derived growth factor plays a key role in proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci 1999;40:2683–2689.
Briggs MC, Grierson I, Hiscott P, et al. Active scatter factor (HGF/SF) in proliferative vitreoretinal disease. Invest Ophthalmol Vis Sci 2000;41:3085–3094.
Ikuno Y, Leong FL, Kazlauskas A . Attenuation of experimental proliferative vitreoretinopathy by inhibiting the platelet-derived growth factor receptor. Invest Ophthalmol Vis Sci 2000;41:3107–3116.
Hinton DR, He S, Jin ML, et al. Novel growth factors involved in the pathogenesis of proliferative vitreoretinopathy. Eye 2002;16:422–428.
Mori K, Gehlbach P, Ando A, et al. Retina-specific expression of PDGF-B versus PDGF-A: vascular versus nonvascular proliferative retinopathy. Invest Ophthalmol Vis Sci 2002;43:2001–2006.
Laqua H, Machemer R . Glial cell proliferation in retinal detachment (massive periretinal proliferation). Am J Ophthalmol 1975;80:602–618.
Van Horn DL, Aaberg TM, Machemer R, et al. Glial cell proliferation in human retinal detachment with massive periretinal proliferation. Am J Ophthalmol 1977;84:383–393.
Nork TM, Wallow IHL, Sramek SJ, et al. Müller's cell involvement in proliferative diabetic retinopathy. Arch Ophthalmol 1987;105:1424–1429.
Fisher SK, Erickson PA, Lewis GP, et al. Intraretinal proliferation induced by retinal detachment. Invest Ophthalmol Vis Sci 1991;32:1739–1748.
Geller SF, Lewis GP, Fisher SK . FGFR1, signaling, and AP-1 expression after retinal detachment: reactive Müller and RPE cells. Invest Ophthalmol Vis Sci 2001;42:1363–1369.
Bringmann A, Reichenbach A . Role of Müller cells in retinal degenerations. Front Biosci 2001;6:E77–E92.
He PM, He S, Garner JA, et al. Retinal pigment epithelial cells secrete and respond to hepatocyte growth factor. Biochem Biophys Res Commun 1998;249:253–257.
Lashkari K, Rahimi N, Kazlauskas A . Hepatocyte growth factor receptor in human RPE cells: implications in proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci 1999;40:149–156.
Shibuki H, Katai N, Kuroiwa S, et al. Expression and neuroprotective effect of hepatocyte growth factor in retinal ischemia-reperfusion injury. Invest Ophthalmol Vis Sci 2002;43:528–536.
Pfaffl M . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:2002–2007.
Limb GA, Salt TE, Munro PM, et al. In vitro characterization of a spontaneously immortalized human Muller cell line (MIO-M1). Invest Ophthalmol Vis Sci 2002;43:864–869.
Machide M, Kamitori K, Kohsaka S . Hepatocyte growth factor-induced differential activation of phospholipase c-gamma 1 and phosphatidylinositol 3-kinase is regulated by tyrosine phosphatase SHP-1 in astrocytes. J Biol Chem 2000;275:31392–31398.
Pournaras CJ . Retinal oxygen distribution: its role in the physiopathology of vasoproliferative microangiopathies. Retina 1995;15:332–347.
Hata Y, Nakagawa K, Ishibashi T, et al. Hypoxia-induced expression of vascular endothelial growth factor by retinal glial cells promotes in vitro angiogenesis. Virchows Arch 1995;426:479–486.
Behzadian MA, Wang XL, Shabrawey M, et al. Effects of hypoxia on glial cell expression of angiogenesis-regulating factors VEGF and TGF-β. Glia 1998;24:216–225.
Eichler W, Kuhrt H, Hoffmann S, et al. VEGF release by retinal glia depends on both oxygen and glucose supply. Neuroreport 2000;11:3533–3537.
Stone J, Maslim J, Valter-Kocsi K, et al. Mechanisms of photoreceptor death and survival in mammalian retina. Prog Retina Eye Res 1999;18:689–735.
Linsenmeier RA, Padnick-Silver L . Metabolic dependence of photoreceptors on the choroid in the normal and detached retina. Invest Ophthalmol Vis Sci 2000;41:3117–3123.
Roque RS, Caldwell RB, Behzadian MA . Cultured Muller cells have high levels of epidermal growth factor receptors. Invest Ophthalmol Vis Sci 1992;33:2587–2595.
D'Amore PA . Mechanisms of retinal and choroidal neovascularization. Invest Ophthalmol Vis Sci 1994;35:3974–3979.
Miller JW, Adamis AP, Shima DT, et al. Vascular endothelial growth factor/vascular permeability factor is temporally and spatially correlated with ocular angiogenesis in a primate model. Am J Pathol 1994;145:574–584.
Frank RN, Amin RH, Eliott D, et al. Basic fibroblast growth factor and vascular endothelial growth factor are present in epiretinal and choroidal neovascular membranes. Am J Ophthalmol 1996;122:393–403.
Milenkovic I, Weick M, Wiedemann P, et al. P2Y receptor-mediated stimulation of Müller glial cell DNA synthesis: dependence on EGF and PDGF receptor transactivation. Invest Ophthalmol Vis Sci 2003;44:1211–1220.
Hollborn M, Jahn K, Limb GA, et al. Characterization of the basic fibroblast growth factor-evoked proliferation of the human Müller cell line, MIO-M1. Graefes Arch Clin Exp Ophthalmol 2004;242, in press.
Williams JA, Sans MD, Tashiro M, et al. Cholecystokinin activates a variety of intracellular signal transduction mechanisms in rodent pancreatic acinar cells. Pharmacol Toxicol 2002;91:297–303.
Yamada T, Tsubouchi H, Daikuhara Y, et al. Immunohistochemistry with antibodies to hepatocyte growth factor and its receptor protein (c-Met) in human brain tissues. Brain Res 1994;637:308–312.
Fenton H, Finch PW, Rubin JS, et al. Hepatocyte growth factor (HGF/SF) in Alzheimer's disease. Brain Res 1998;779:262–270.
Hirose Y, Kojima M, Sagoh M, et al. Immunohistochemical examination of c-Met protein expression in astrocytic tumors. Acta Neuropathol (Berl) 1998;95:345–351.
Nabeshima K, Shimao Y, Sato S, et al. Expression of c-Met correlates with grade of malignancy in human astrocytic tumours: an immunohistochemical study. Histopathology 1997;31:436–443.
Koochekpour S, Jeffers M, Rulong S, et al. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res 1997;57:5391–5398.
Welch WC, Kornblith PL, Michalopoulos GK, et al. Hepatocyte growth factor (HGF) and receptor (c-met) in normal and malignant astrocytic cells. Anticancer Res 1999;19:1635–1640.
Acknowledgements
This work was supported in part by the Bundesministerium für Bildung, Forschung und Technologie (Interdisciplinary Center for Clinical Research at the University of Leipzig, 01KS9504, Project C21), and by the Deutsche Forschungsgemeinschaft (Ko1547/4-1, and Br 1249/2-1). We thank Mrs K Bartholomäus, U Weinbrecht, and F Kutzera for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hollborn, M., Krausse, C., Iandiev, I. et al. Glial cell expression of hepatocyte growth factor in vitreoretinal proliferative disease. Lab Invest 84, 963–972 (2004). https://doi.org/10.1038/labinvest.3700121
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700121
Keywords
This article is cited by
-
Stage specificity of novel growth factor expression during development of proliferative vitreoretinopathy
Eye (2007)
-
Albumin marks pseudopodia of astrocytoma cells responding to hepatocyte growth factor or serum
Laboratory Investigation (2006)
-
Proteomic characterization of harvested pseudopodia with differential gel electrophoresis and specific antibodies
Laboratory Investigation (2005)