Featured
-
-
Article
| Open Access Reconstructing metastatic seeding patterns of human cancers
Reconstructing metastatic seeding patterns of human cancers
Tumours frequently metastasize to multiple anatomical sites and understanding how these different metastases evolve may be important for therapy. Here, the authors develop a method—Treeomics—that can construct phylogenies from multiple metastases from next-generation sequencing data.
- Johannes G. Reiter
- , Alvin P. Makohon-Moore
- & Martin A. Nowak
-
Article
| Open Access Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast
Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast
Fission yeastSchizosaccharomyces pombe has diverse traits. Jeffares et al. characterize large copy number variations (CNVs) and rearrangements in S. pombe, and show that CNVs are transient with effects on quantitative traits and gene expression, whereas rearrangements influence intrinsic reproductive isolation.
- Daniel C. Jeffares
- , Clemency Jolly
- & Fritz J. Sedlazeck
-
Article
| Open Access A cell-based computational model of early embryogenesis coupling mechanical behaviour and gene regulation
A cell-based computational model of early embryogenesis coupling mechanical behaviour and gene regulation
Embryonic development is a complex process where genetic and biochemical information direct morphogenesis. Here the authors describe MecaGen, an agent-based model and simulation platform of multicellular development designed to allow a quantitative comparison between simulations and real biological data.
- Julien Delile
- , Matthieu Herrmann
- & René Doursat
-
Article
| Open Access MetaSort untangles metagenome assembly by reducing microbial community complexity
MetaSort untangles metagenome assembly by reducing microbial community complexity
Currently available metagenomic data analysis relies on reference genomes. Here, the authors describe a newde novometagenomic assembly method, metaSort, that constructs bacterial genomes from metagenomic samples to reduce microbial community complexity while increasing genome recovery and assembly.
- Peifeng Ji
- , Yanming Zhang
- & Fangqing Zhao
-
Article
| Open Access Rapid tuning shifts in human auditory cortex enhance speech intelligibility
Rapid tuning shifts in human auditory cortex enhance speech intelligibility
Experience constantly shapes perception, but the neural mechanisms of this rapid plasticity are unclear. Here, Holdgraf et al. record neural activity in the human auditory cortex and show that listening to normal speech elicits rapid plasticity that increases the neural gain for features of sound that are key for speech intelligibility.
- Christopher R. Holdgraf
- , Wendy de Heer
- & Frédéric E. Theunissen
-
Article
| Open Access Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism
Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism
Altered DNA methylation is a feature of cancer and between-patient variability is prevalent. Here, the authors integrate data on thousands of human tumours, and find that expression levels of methionine metabolism genes are predictive of methylation features, and that the breakdown of this relationship is a negative prognostic marker.
- Mahya Mehrmohamadi
- , Lucas K. Mentch
- & Jason W. Locasale
-
Article
| Open Access Defining functional interactions during biogenesis of epithelial junctions
Defining functional interactions during biogenesis of epithelial junctions
Formation and reinforcement of E-cadherin-mediated adhesion depends on intracellular trafficking and interactions with the actin cytoskeleton, but how these are coordinated is not known. Here the authors conduct a focused phenotypic screen to identify new pathways regulating cell–cell junction homeostasis.
- J. C. Erasmus
- , S. Bruche
- & V. M. M. Braga
-
Article
| Open Access An ethnically relevant consensus Korean reference genome is a step towards personal reference genomes
An ethnically relevant consensus Korean reference genome is a step towards personal reference genomes
The utility of a universal reference sequence for human genome comparisons is dependent on the ethnic origins of the individuals being sequenced. Here the authors report a Korean reference genome and consensus variome, and show that an ethnically-relevant reference can improve variant detection.
- Yun Sung Cho
- , Hyunho Kim
- & Jong Bhak
-
Article
| Open Access A principal component meta-analysis on multiple anthropometric traits identifies novel loci for body shape
A principal component meta-analysis on multiple anthropometric traits identifies novel loci for body shape
Past genome-wide associate studies have identified hundreds of genetic loci that influence body size and shape when examined one trait at a time. Here, Jeff and colleagues develop an aggregate score of various body traits, and use meta-analysis to find new loci linked to body shape.
- Janina S. Ried
- , Janina Jeff M.
- & Ruth J. F. Loos
-
Article
| Open Access Deep phenotyping unveils hidden traits and genetic relations in subtle mutants
Deep phenotyping unveils hidden traits and genetic relations in subtle mutants
Experimenter scoring of cellular imaging data can be biased. This study describes an automated and unbiased multidimensional phenotyping method that relies on machine learning and complex feature computation of imaging data, and identifies weak alleles affecting synapse morphology in live C. elegans.
- Adriana San-Miguel
- , Peri T. Kurshan
- & Hang Lu
-
Article
| Open Access A systems study reveals concurrent activation of AMPK and mTOR by amino acids
A systems study reveals concurrent activation of AMPK and mTOR by amino acids
mTORC1 is known to mediate the signalling activity of amino acids. Here, the authors combine modelling with experiments and find that amino acids acutely stimulate mTORC2, IRS/PI3K and AMPK, independently of mTORC1. AMPK activation through CaMKKβ sustains autophagy under non-starvation conditions.
- Piero Dalle Pezze
- , Stefanie Ruf
- & Kathrin Thedieck
-
Article
| Open Access De-novo protein function prediction using DNA binding and RNA binding proteins as a test case
De-novo protein function prediction using DNA binding and RNA binding proteins as a test case
Identification of the function of proteins is difficult when there are no structurally or biochemically characterized homologs. Here, the authors present an approach that allows the prediction of nucleic-acid binding proteins based on sequence alone, and they are able to experimentally validate their method.
- Sapir Peled
- , Olga Leiderman
- & Yanay Ofran
-
Article
| Open Access Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst
Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst
Early embryonic cell fate and lineage specification is tightly regulated in the preimplantation mammalian embryo. Here, the authors quantitatively examine the ratio of epiblast to primitive endoderm lineages in the blastocyst and show composition of the inner cell mass is conserved, independent of its size.
- Néstor Saiz
- , Kiah M. Williams
- & Anna-Katerina Hadjantonakis
-
Article
| Open Access Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins
Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins
Protein repeats may be considered a paradox, being evolutionarily conserved yet also hotspots of protein evolution associated with innovation. Here, the authors use a novel method to show that new repeats undergo rapid divergence within species, but are then fixed and conserved between species.
- Erez Persi
- , Yuri I. Wolf
- & Eugene V Koonin
-
Article
| Open Access In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development
In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development
Pathway analysis aids interpretation of large-scale gene expression data, but existing algorithms fall short of providing robust pathway identification. The method introduced here includes coexpression analysis and gene importance estimation to robustly identify relevant pathways and biomarkers for patient stratification.
- Ivan V. Ozerov
- , Ksenia V. Lezhnina
- & Alex Zhavoronkov
-
Article
| Open Access A multi-marker association method for genome-wide association studies without the need for population structure correction
A multi-marker association method for genome-wide association studies without the need for population structure correction
Currently available methods for phenotype to genetic markers association need to account for population structure. Here, Klasen et al. devise a statistical method called Quantitative Trait Cluster Association Test (QTCAT) that overcomes the need for population structure correction.
- Jonas R. Klasen
- , Elke Barbez
- & Korbinian Schneeberger
-
Article
| Open Access Rapid construction of a whole-genome transposon insertion collection for Shewanella oneidensis by Knockout Sudoku
Rapid construction of a whole-genome transposon insertion collection for Shewanella oneidensis by Knockout Sudoku
Knockout collections provide a valuable tool to explore gene function, yet are expensive and technically challenging to produce at a genome-wide scale. Here Baym et al. devise a cost-effective transposon-based method to quickly develop a knockout collection for the electroactive microbe Shewanella oneidensis.
- Michael Baym
- , Lev Shaket
- & Buz Barstow
-
Article
| Open Access MHC class II complexes sample intermediate states along the peptide exchange pathway
MHC class II complexes sample intermediate states along the peptide exchange pathway
MHCII proteins bind and present both foreign and self-antigens to potentially activate CD4+ T cells via cognate T cell receptors (TCRs) during the adaptive immune response. Here, the authors combine NMR-detected H/D exchange with Markov modelling analysis to shed light on the dynamics of MHCII peptide exchange.
- Marek Wieczorek
- , Jana Sticht
- & Christian Freund
-
Article
| Open Access Random synaptic feedback weights support error backpropagation for deep learning
Random synaptic feedback weights support error backpropagation for deep learning
Multi-layered neural architectures that implement learning require elaborate mechanisms for symmetric backpropagation of errors that are biologically implausible. Here the authors propose a simple resolution to this problem of blame assignment that works even with feedback using random synaptic weights.
- Timothy P. Lillicrap
- , Daniel Cownden
- & Colin J. Akerman
-
Article
| Open Access Quantifying unobserved protein-coding variants in human populations provides a roadmap for large-scale sequencing projects
Quantifying unobserved protein-coding variants in human populations provides a roadmap for large-scale sequencing projects
Accurate estimations of the frequency distribution of rare variants are needed to quantify the discovery power and guide large-scale human sequencing projects. This study describes an algorithm called UnseenEst to estimate the distribution of genetic variations using tens of thousands of exomes.
- James Zou
- , Gregory Valiant
- & Daniel G. MacArthur
-
Article
| Open Access Self-motion evokes precise spike timing in the primate vestibular system
Self-motion evokes precise spike timing in the primate vestibular system
Early vestibular pathways are thought to code sensory inputs regarding self-motion via changes in firing rate. Here, the authors record from both regular and irregular afferents in macaques, and find both irregular afferents and central neurons also represent self-motion via temporally precise spike timing.
- Mohsen Jamali
- , Maurice J. Chacron
- & Kathleen E. Cullen
-
Article
| Open Access Pan-cancer transcriptomic analysis associates long non-coding RNAs with key mutational driver events
Pan-cancer transcriptomic analysis associates long non-coding RNAs with key mutational driver events
Long non-coding RNAs are implicated in multiple aspects of tumourigenesis. Here, the authors generate a landscape of these macromolecules in a wide array of cancer types and examine which RNAs are transcriptionally altered in relation to somatic driver mutations in established coding cancer genes.
- Arghavan Ashouri
- , Volkan I. Sayin
- & Erik Larsson
-
Article
| Open Access A network property necessary for concentration robustness
A network property necessary for concentration robustness
Absolute concentration robustness (ACR), independence of the steady-state concentration of a molecule from the environment, is difficult to predict. Here, the authors derive a network structure-based necessary condition for ACR, and suggest that metabolites satisfying the condition are prevalent.
- Jeanne M. O. Eloundou-Mbebi
- , Anika Küken
- & Zoran Nikoloski
-
Article
| Open Access Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival
Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival
Cancer cells reprogramme their metabolism with unclear clinical implications. Here, the authors analyse the expression of metabolic genes across 20 types of solid cancers and find that clinical aggressiveness, poor survival and metastasis are associated with the deregulation of mitochondrial metabolism.
- Edoardo Gaude
- & Christian Frezza
-
Article
| Open Access Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli
Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli
Multi-omics data integration is a great challenge. Here, the authors compile a database of E. coliproteomics, transcriptomics, metabolomics and fluxomics data to train models of recurrent neural network and constrained regression, enabling prediction of bacterial responses to perturbations.
- Minseung Kim
- , Navneet Rai
- & Ilias Tagkopoulos
-
Article
| Open Access A high-quality human reference panel reveals the complexity and distribution of genomic structural variants
A high-quality human reference panel reveals the complexity and distribution of genomic structural variants
Structural variants (SVs) are prevalent in genomes of the general population. Here, Guryev and The Genome of the Netherlands Consortium describe the reference panel of haplotype-resolved SVs from 769 individuals from 250 Dutch families and show its utility for studying heritable traits.
- Jayne Y. Hehir-Kwa
- , Tobias Marschall
- & Victor Guryev
-
Article
| Open Access Modelling proteins’ hidden conformations to predict antibiotic resistance
Modelling proteins’ hidden conformations to predict antibiotic resistance
Expression of TEM β-lactamase is a predominant mechanism underlying antibiotic resistance in pathogenic Gram-negative bacteria. Here, the authors use Markov state models to reveal and experimentally confirm hidden conformations that determine TEM substrate specificity.
- Kathryn M. Hart
- , Chris M. W. Ho
- & Gregory R. Bowman
-
Article
| Open Access Comparative survey of the relative impact of mRNA features on local ribosome profiling read density
Comparative survey of the relative impact of mRNA features on local ribosome profiling read density
Ribosome profiling data can suffer from uneven coverage which hampers estimation of elongation rates. Connor et al.present an enhanced data smoothing method for Ribo-seq data and highlight significant variability in sequence determinants of ribosome density in publicly available data sets.
- Patrick B. F. O’Connor
- , Dmitry E. Andreev
- & Pavel V. Baranov
-
Article
| Open Access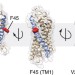 Dimerization deficiency of enigmatic retinitis pigmentosa-linked rhodopsin mutants
Dimerization deficiency of enigmatic retinitis pigmentosa-linked rhodopsin mutants
Retinitis pigmentosa is often caused by mutations that affect the activity or transport of rhodopsin, but some mutations cause disease even though an apparently functional protein is produced. Here the authors show that three such enigmatic mutants retain scramblase activity but are unable to dimerize.
- Birgit Ploier
- , Lydia N. Caro
- & Anant K. Menon
-
Article
| Open Access Dichotomy of cellular inhibition by small-molecule inhibitors revealed by single-cell analysis
Dichotomy of cellular inhibition by small-molecule inhibitors revealed by single-cell analysis
Many drugs are small molecule inhibitors of cell signalling. Through single cell analysis and mathematical modelling here the authors show that cell-to-cell variability diversifies inhibition response into digital and analogue, and that the two translate into distinct long-term functional responses.
- Robert M. Vogel
- , Amir Erez
- & Grégoire Altan-Bonnet
-
Article
| Open Access Forward design of a complex enzyme cascade reaction
Forward design of a complex enzyme cascade reaction
Building multi-component enzymatic processes in one pot is challenged by the inherent complexity of each biochemical system. Here, the authors use online mass spectroscopy and engineering systems theory to achieve forward design of a ten-membered reaction cascade.
- Christoph Hold
- , Sonja Billerbeck
- & Sven Panke
-
Article
| Open Access Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd
Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd
A wealth of gene expression data is publicly available, yet is little use without additional human curation. Ma’ayan and colleagues report a crowdsourcing project involving over 70 participants to annotate and analyse thousands of human disease-related gene expression datasets.
- Zichen Wang
- , Caroline D. Monteiro
- & Avi Ma’ayan
-
Article
| Open Access Comparative genomics reveals adaptive evolution of Asian tapeworm in switching to a new intermediate host
Comparative genomics reveals adaptive evolution of Asian tapeworm in switching to a new intermediate host
Only one of the three Taenia species causing taeniasis in humans was previously sequenced. Here the authors provide draft genomes of Taenia saginata and Taenia asiatica, analyse genome evolution of all three species, and identify potential targets for developing diagnostic markers or intervention tools.
- Shuai Wang
- , Sen Wang
- & Xuepeng Cai
-
Article
| Open Access Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes
Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes
Plasticity and clonal population structure in bacterial genomes can hinder traditional SNP-based genetic association studies. Here, Corander and colleagues present a method to identify variable-length sequence elements enriched in a phenotype of interest, and demonstrate its use in human pathogens.
- John A. Lees
- , Minna Vehkala
- & Jukka Corander
-
Article
| Open Access Rare variant phasing and haplotypic expression from RNA sequencing with phASER
Rare variant phasing and haplotypic expression from RNA sequencing with phASER
Genome interpretation and analysis of allelic activity requires appropriate haplotype phasing. Here the authors present phASER, a fast and accurate method for variant phrasing from RNA-seq and genome sequencing data.
- Stephane E. Castel
- , Pejman Mohammadi
- & Tuuli Lappalainen
-
Article
| Open Access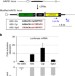 Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity
Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity
To modulate gene expression, the glucocorticoid receptor binds to response elements (RE) that vary in sequence. Here, the authors show that RE sequences can modulate glucocorticoid receptor structure and activity, which might provide regulatory specificity towards individual target genes.
- Stefanie Schöne
- , Marcel Jurk
- & Sebastiaan H. Meijsing
-
Article
| Open Access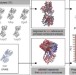 Prediction and validation of protein intermediate states from structurally rich ensembles and coarse-grained simulations
Prediction and validation of protein intermediate states from structurally rich ensembles and coarse-grained simulations
Protein conformational changes are key to a wide range of cellular functions but remain difficult to access experimentally. Here the authors describe eBDIMS, a novel approach to predict intermediates observed in structural transition pathways from experimental ensembles.
- Laura Orellana
- , Ozge Yoluk
- & Erik Lindahl
-
Article
| Open Access Prediction of allosteric sites and mediating interactions through bond-to-bond propensities
Prediction of allosteric sites and mediating interactions through bond-to-bond propensities
Allostery is a key molecular mechanism underpinning control and modulation in a variety of cellular processes. Here, the authors present a method that can be used to predict allosteric sites and the mediating interactions that connect them to the active site of the protein.
- B. R. C. Amor
- , M. T. Schaub
- & M. Barahona
-
Article
| Open Access Crowdsourced assessment of common genetic contribution to predicting anti-TNF treatment response in rheumatoid arthritis
Crowdsourced assessment of common genetic contribution to predicting anti-TNF treatment response in rheumatoid arthritis
Rheumatoid arthritis patients respond differently to anti-TNF treatment. Using community-based challenge, the authors show that currently available data does not reveal meaningful genetic predictors of response to anti-TNF therapy, thus confirming clinical observations.
- Solveig K. Sieberts
- , Fan Zhu
- & Lara M. Mangravite
-
Article
| Open Access Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults
Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults
Clonal haematopoiesis has been thought to occur in less than 10% of individuals younger than 70 years old. Here, the authors use an error corrected next-generation sequencing method to find clonal haematopoiesis in the peripheral blood of 19 of 20 healthy 50–70 year old individuals.
- Andrew L. Young
- , Grant A. Challen
- & Todd E. Druley
-
Article
| Open Access reChIP-seq reveals widespread bivalency of H3K4me3 and H3K27me3 in CD4+ memory T cells
reChIP-seq reveals widespread bivalency of H3K4me3 and H3K27me3 in CD4+ memory T cells
Co-localizing chromatin modifications and regulators can exert a combinatorial effect on chromatin structure and function. Here the authors describe reChIP-seq and normR to identify co-localizing proteins in an unbiased genome-wide manner.
- Sarah Kinkley
- , Johannes Helmuth
- & Ho-Ryun Chung
-
Article
| Open Access Extension of human lncRNA transcripts by RACE coupled with long-read high-throughput sequencing (RACE-Seq)
Extension of human lncRNA transcripts by RACE coupled with long-read high-throughput sequencing (RACE-Seq)
Long non-coding RNAs are increasingly recognised to be important factors in regulating cellular processes and comprise a large faction of the transcriptome, however most are uncharacterised. Here the authors present RACE-Seq, a tool to improve and extend the annotation of low-expression transcripts.
- Julien Lagarde
- , Barbara Uszczynska-Ratajczak
- & Jennifer Harrow
-
Article
| Open Access Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features
Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features
Diagnosis of lung cancer through manual histopathology evaluation is insufficient to predict patient survival. Here, the authors use computerized image processing to identify diagnostically relevant image features and use these features to distinguish lung cancer patients with different prognoses.
- Kun-Hsing Yu
- , Ce Zhang
- & Michael Snyder
-
Article
| Open Access Environmental change makes robust ecological networks fragile
Environmental change makes robust ecological networks fragile
Despite their complexity, ecological networks appear robust to species loss. Here, Strona and Lafferty use artificial life simulations and real-world data to show that such robustness applies to stable conditions, but can collapse when the environment changes.
- Giovanni Strona
- & Kevin D. Lafferty
-
Article
| Open Access Self-organized centripetal movement of corneal epithelium in the absence of external cues
Self-organized centripetal movement of corneal epithelium in the absence of external cues
The cornea is formed of cells that originate from the outer circle of stem cells and that move towards its centre. Here, the authors show that the movement pattern is self-organised, requiring no cues, and that stem cell leakage may account for the presence of stem cells at the centre of the cornea.
- Erwin P. Lobo
- , Naomi C. Delic
- & J. Guy Lyons
-
Article
| Open Access High-resolution imaging and computational analysis of haematopoietic cell dynamics in vivo
High-resolution imaging and computational analysis of haematopoietic cell dynamics in vivo
It is difficult to image haematopoietic stem cells (HSC) in their niche. Here, the authors present a new high-throughput computational approach to visualise HSCs in vivoat a high spatial and temporal resolution and also use a Msi2-reporter to label endogenous HSCs and progenitors, enabling cell tracking
- Claire S. Koechlein
- , Jeffrey R. Harris
- & Tannishtha Reya
-
Article
| Open Access Analysis of chromosomal aberrations and recombination by allelic bias in RNA-Seq
Analysis of chromosomal aberrations and recombination by allelic bias in RNA-Seq
Chromosomal aberrations can be detected by global gene expression analysis. Here, the authors report eSNP-Karyotyping, a new method that can detect chromosomal aberrations by measuring the ratio of expression between the two alleles without comparison to a matched diploid sample.
- Uri Weissbein
- , Maya Schachter
- & Nissim Benvenisty
-
Article
| Open Access Mpath maps multi-branching single-cell trajectories revealing progenitor cell progression during development
Mpath maps multi-branching single-cell trajectories revealing progenitor cell progression during development
The tools for constructing cell lineages from single cell data are limited. Here, the authors present Mpath: an algorithm to derive multi-branching trajectories using neighbourhood-based cell state transitions, and use this to predict developmental trajectories in dendritic and muscle cells.
- Jinmiao Chen
- , Andreas Schlitzer
- & Michael Poidinger
-
Article
| Open Access Information recovery from low coverage whole-genome bisulfite sequencing
Information recovery from low coverage whole-genome bisulfite sequencing
Here, Libertini and colleagues devise a computation tool that can analyze whole-genome bisulfite sequencing (WGBS) data to recover of ∼30% of the lost differential methylation position information. They use COMETgazer and COMETvintage to analyze 13 diffferent methylome data to demonstrate their performance.
- Emanuele Libertini
- , Simon C. Heath
- & Stephan Beck
Browse broader subjects
Browse narrower subjects
- Biochemical reaction networks
- Cellular signalling networks
- Classification and taxonomy
- Communication and replication
- Computational models
- Computational neuroscience
- Computational platforms and environments
- Data acquisition
- Data integration
- Data mining
- Data processing
- Data publication and archiving
- Databases
- Functional clustering
- Gene ontology
- Gene regulatory networks
- Genome informatics
- Hardware and infrastructure
- High-throughput screening
- Image processing
- Literature mining
- Machine learning
- Microarrays
- Network topology
- Phylogeny
- Power law
- Predictive medicine
- Probabilistic data networks
- Programming language
- Protein analysis
- Protein design
- Protein folding
- Protein function predictions
- Protein structure predictions
- Proteome informatics
- Quality control
- Scale invariance
- Sequence annotation
- Software
- Standards
- Statistical methods
- Virtual drug screening

 Pancancer modelling predicts the context-specific impact of somatic mutations on transcriptional programs
Pancancer modelling predicts the context-specific impact of somatic mutations on transcriptional programs