Abstract
The expression of adenovirus late genes is shown to require viral DNA replication, but its mechanism remains elusive. Here we found that knockdown of CTCF suppresses viral DNA replication as well as late, but not early, gene expression. Chromatin immunoprecipitation assays indicated that CTCF binds to viral chromatin depending on viral DNA replication. These findings depict CTCF as a critical regulator for adenovirus genome functions in late phases of infection.
Similar content being viewed by others
Introduction
In the cell nucleus, the genomic DNA forms chromatin structure. It is being clarified that the higher-order chromatin structure, such as the DNA looping, plays an important role in a dynamic property of the chromatin1. One of the chromatin organizing proteins, CTCF (CCCTC-binding factor), is a well-characterized chromatin-binding factor involved in the formation of the long-range interactions of chromatin2. CTCF has eleven zinc fingers and therefore binds to divergent DNA sequences, as indicated by chromatin immunoprecipitation (ChIP) in combination with tiling arrays (ChIP-on-chip)3 or high-throughput sequencing analyses (ChIP-seq)4. A variety of chromatin-related proteins are reported as binding partners of CTCF, including cohesin complexes5,6, a nucleolar protein B23/nucleophosmin and CTCF itself 7. These interactions are thought to enable CTCF binding sites to contact each other and/or be tethered to the subnuclear domains, resulting in the formation of intra- and interchromatin interaction2. In addition to the role on the cellular chromatin, recent reports have revealed the involvement of CTCF on viral proliferation, as Lieberman and co-workers recently demonstrated the CTCF-mediated formation of chromatin loops on Kaposi's sarcoma-associated Herpesvirus (KSHV) and Epstein-Barr virus (EBV) genomes8,9. It is shown that CTCF regulates the latency-specific chromatin conformation of KSHV and EBV genomes and siRNA-mediated depletion of CTCF or mutations in the CTCF binding sites disrupt the chromatin architecture and de-regulate latent gene expression8,9. Thus, CTCF could impact on the regulation of not only cellular but also viral chromatin.
The adenovirus (Ad) has a linear double-stranded DNA genome that forms chromatin-like structure in the virion10. Previously, we have reported that viral chromatin structure regulates the expression of viral early genes (e.g. E1A, E4 genes) in early phases of infection11,12. The expression of the late genes (e.g. major late genes) are hardly observed during early phases of infection, while concomitantly with the onset of viral DNA replication, those genes are fully activated. Thomas and Mathews demonstrated that the expression of the late genes requires viral DNA replication in cis13. In addition, we have shown the regulatory mechanism of the viral chromatin structure during DNA replication and proposed a possible role of viral DNA replication in the activation of late genes14. Thus, it is suggested that the regulation of viral chromatin structure has a significant role in the DNA replication-dependent activation of viral genes. In spite of these evidences, however, the functional relationship between viral gene expression and DNA replication in infected cells remains largely unclear. In this study we sought to further clarify the role of chromatin structure and/or chromatin-related factors on the Ad genome DNA. As described above, it is shown that CTCF plays a role on the chromatin of some DNA viruses8,9. These lead us to hypothesize that CTCF could function also on Ad chromatin.
Results
CTCF is required for viral DNA replication and late gene expression
To study a role of CTCF, we carried out knock down (KD) of the expression of CTCF by siRNA treatment (Fig. 1A). Either control siRNA (siCont) or siRNA targeted for CTCF (siCTCF) was introduced into HeLa cells and then cell lysates were prepared and subjected to western blot analyses using anti-CTCF antibody. Only a single band corresponding to CTCF was detected, demonstrating the specificity of the antibody. Under our experimental condition, the expression level of CTCF in siCTCF-treated cells was decreased to approximately 25% of that in control cells (Fig. 1A, compare lane 4 with lanes 1–3).
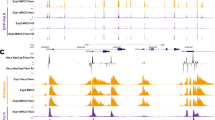
Effects of CTCF KD on viral DNA replication and gene expression.
(A) Western blotting and knockdown of CTCF. Cell lysates were prepared from HeLa cells treated with siCont (lanes 3) or siCTCF (lane 4) and subjected to western blot analyses using anti-CTCF (top panel) and anti-β-actin antibodies (bottom panel). For siCont-treated cells, 25% (lane 1) and 50% (lane 2) volume of lysates were also loaded. Full-size images are shown in Supplementary figure 1. (B) Amounts of viral DNA. HeLa cells treated with siCont or siCTCF were infected with HAdV5 at an MOI of 100 and total DNAs were purified at 6 and 12 hpi. The amount of viral DNA was measured by qPCR using primers for the E1A promoter. The amount at 12 hpi relative to that at 6 hpi was graphed. Mean values with s.d. were obtained from three independent experiments. (C) RT-qPCR assays. Total RNAs were purified at 6 and 12 hpi and subjected to RT-qPCR using indicated primer sets. The mRNA levels relative to those of control cells at 12 hpi were graphed. Mean values with s.d. were obtained from three independent experiments. P-values are calculated using Student's t-test. “n.s.” indicates “not statistically significant”.
To test whether CTCF plays a role in Ad DNA replication and gene expression, we carried out CTCF KD followed by quantitative PCR (qPCR) of viral DNA and RT-qPCR (Fig. 1B and C). Under our experimental condition, the onset of viral DNA replication can be observed around 8 hpi (hours post infection)12,14. siCont- or siCTCF-treated cells were mock-infected or infected with human adenovirus type 5 (HAdV5) at an MOI (multiplicity of infection) of 100 and at 6 (for early phases) and 12 hpi (for late phases of infection) total DNAs and RNAs were purified. We first measured viral DNA amounts by qPCR using a primer set targeted for the E1A promoter region (E1A pro, see 1Table 2) to evaluate the efficiency of viral DNA replication (Fig. 1B). In siCont-treated cells, the amount of viral DNA was increased by ~30 fold through viral DNA replication. In contrast, siCTCF-treated cells allowed only ~9 fold amplification of viral DNA.
Next, we performed RT-qPCR assays using several primer sets for cellular and viral genes (Fig. 1C). Under the condition employed here, the mRNA level of GAPDH was unaffected by Ad infection and siRNA treatment and that of CTCF was specifically decreased by siCTCF treatment (Fig. 1C, GAPDH and CTCF). The mRNA levels of viral early genes were not significantly affected by CTCF KD (Fig. 1C, E1A and E4), suggesting that CTCF is not involved in viral early gene expression. In contrast, the level of mRNA transcribed from the major late promoter (MLP) was drastically decreased by siCTCF treatment (Fig. 1C, MLP). Similarly, the mRNA level of E2A was reduced by CTCF KD, particularly at 12 hpi (Fig. 1C, E2A). It is noted that E2 gene transcription is regulated by early and late promoters and transcription from the E2 late promoter depends on viral DNA replication15 (Unpublished data). Therefore, it is reasonable to assume that CTCF KD could predominantly affect the transcription from the E2 late promoter, although in this study we did not discriminate E2A mRNAs transcribed from two promoters. Collectively, these results suggest that CTCF is critical for viral DNA replication as well as late, but not early, gene expression.
CTCF binds to viral chromatin in a DNA replication-dependent manner
To examine whether CTCF functions directly on viral chromatin, ChIP assays were performed using anti-CTCF antibody (Fig. 2). Since the effect of CTCF KD was observed in late phases of infection (Fig. 1), first we studied using infected cells at 12 hpi for ChIP assays (Fig. 2A). We used a variety of primer sets for the Ad genome to test the genome-wide binding of CTCF (see Fig. 2B). We found that CTCF is recruited into several regions of the virus genome, including the MLP region and the ORF regions of the viral structural protein (Hexon) and the E4 ORF3 gene (E4 orf). In addition, a weak binding of CTCF at the E1A pro region was observed. Next, we focused on three CTCF binding sites, the E1A pro, MLP and Hexon regions and performed ChIP assays using cells cultured in the absence or presence of hydroxyurea (HU), a DNA replication inhibitor, to examine whether the CTCF binding observed here is DNA replication-dependent (Fig. 2C). The recruitment of CTCF into those regions was observed only at 12 hpi and this was inhibited by the addition of HU, indicating that CTCF is recruited onto viral chromatin in a DNA replication-dependent manner.
CTCF binding on viral chromatin.
(A) ChIP assays with anti-CTCF antibody. HeLa cells were infected with HAdV5 at an MOI of 100 and at 12 hpi subjected to ChIP assays using anti-CTCF and anti-FLAG antibodies. The binding levels were calculated as fold enrichment against that obtained in a negative control (anti-FLAG antibody). Mean values with s.d. were obtained from three independent experiments. (B) Structure of the Ad genome. Arrows represent promoters of viral genes. Target regions for ChIP assays are indicated by gray bars. (C) Effect of viral DNA replication on CTCF binding. HeLa cells were infected with HAdV5 and cultured in the absence or presence of 2 mM HU. At 6 and 12 hpi, ChIP assays were carried out as described above. P-values are calculated using Student's t-test.
Discussion
The results obtained in this study indicate that CTCF binds to Ad chromatin depending on its DNA replication and plays a pivotal role in late phases of infection. Our KD experiments clearly reveal that CTCF is required for viral DNA replication and late gene expression (Fig. 1B and C). It is demonstrated that viral late genes are activated depending on its DNA replication13. Thus, the mRNA levels of late genes also should be affected when only DNA replication would be directly repressed by CTCF KD. Conversely, viral factors involved in its DNA replication are encoded by the E2 gene and its expression was suppressed by CTCF KD (Fig. 1C). Therefore, the inhibition of viral late gene expression also could be the cause for less efficient viral DNA replication. Because of this interdependency, we could not precisely discriminate whether CTCF KD primarily affects viral DNA replication or late gene expression (or both). In addition, we could not exclude the possibility that the lower level of the MLP mRNA in siCTCF-treated cells results from less amount of viral DNA templates due to the defect in viral DNA replication. Nonetheless, we speculate that CTCF may be involved in the regulation of late gene expression: First, the binding of CTCF is dependent on viral DNA replication (Fig. 2C), suggesting that this protein possibly functions in the step(s) later than viral DNA replication; second, when the level of the MLP mRNA at 12 hpi was normalized by the amounts of the virus genome, the level of the MLP mRNA per one viral DNA in CTCF KD cells was still lower than that in control cells (Fig. 1, MLP mRNA level: ~15%/virus genome: ~30% = MLP mRNA per one viral DNA: ~50%).
It is an important question how CTCF regulates the function of the virus genome/chromatin. Although in this study we found several CTCF binding sites (Fig. 2A), there could be additional regions for CTCF on the virus genome. Thus, at the moment, it is difficult to dissect a role of each CTCF binding site and the cooperative function among those regions. It is suggested that the E1B, IX and E2 late genes/promoters are also activated depending on viral DNA replication15,16,17 as is the MLP. However, we could not observe the CTCF binding on those regions (Fig. 2A), suggesting that the recruitment of CTCF onto each late promoter seems not to be required for the genome-wide coordination between viral DNA replication and the expression of late genes. Further studies are needed to address these points. As described above, it is shown that Ad DNA replication is required for the activation of viral late genes13. Similarly it is reported that DNA replication is essential for the expression of certain cellular genes such as the HoxB gene18, although the molecular details remain to be determined. To our knowledge, this is the first report indicating the possible involvement of CTCF in the DNA replication-dependent activation of the genes. Thus, our findings may provide insight into an uncharacterized mechanism of gene regulation that involves DNA replication.
Methods
Cells and viruses
Maintenance of HeLa cells and purification and infection of human adenovirus type 5 (HAdV5) were carried out essentially as described previously12,14. Hydroxyurea (HU) was added at the final concentration of 2 mM right after infection to block DNA replication.
Antibodies
To obtain recombinant CTCF N-terminal region (amino acids (aa) 1–267) as an antigen, the expression vector for His-tagged CTCF(1–267) was constructed. cDNA fragment of full-length CTCF was amplified by PCR with a primer set, 5′-AGGGCATATGGAAGGTGATGCAGTCGAAGCCATTGTGG-3' and 5′-AGCCTCGAGAAGTCCTGGCGACGCACAAGGCTCCGCC-3' and cloned into the pBluescript-FLAG vector (pBS-FLAG-CTCF). Using pBS-FLAG-CTCF as a template, cDNA fragment corresponding to aa 1-267 was amplified by PCR with a primer set, 5′-AGGGCATATGGAAGGTGATGCAGTCGAAGCCATTGTGG-3' and 5′-GTTGAATTCACTGGAATGTCTTCTTTACAC-3′ and cloned into the pET-14b vector. E. coli was transformed with the resultant vector, pET-14b-CTCF(1-267) and His-CTCF(1–267) was expressed and purified using the Ni-NTA resin (Novagen) according to the manufacturer's protocol.
Rabbit anti-CTCF antibody was raised against His-CTCF(1–267) according to standard protocols. Mouse anti-FLAG M2 and mouse anti-β-actin antibodies were described elsewhere12,14.
RT-qPCR assays
RT-PCR and quantitative PCR (qPCR) were performed essentially as described previously12,14. Total RNAs were purified by phenol extraction followed by DNase I treatment. cDNA was synthesized from total RNA (1 μg) using ReverTraAce (Toyobo) and oligo-dT primer according to the manufacturer's protocol. qPCR was carried out using FastStart SYBR Green Master (Roche) and Thermal Cycler Dice Real Time System (Takara) according to the manufacturers' protocol.
Table 1 indicates primer sequences for GAPDH, CTCF, E1A, E4, MLP and E2A genes.
ChIP assays
ChIP assays were carried out according to the protocol from Chromatin Immunoprecipitation Assay Kit (Millipore) with minor modification, essentially as described previously12,14. Briefly, cells were fixed with 1% formaldehyde, followed by the addition of glycine at the final concentration of 125 mM for quenching. After centrifugation, cell pellets were lysed with SDS lysis buffer (50 mM Tris-HCl [pH 7.9], 10 mM EDTA and 1% SDS) and lysates were subjected to sonication to shear the chromatin DNA to ~1 kbp in size. Sonicated samples were diluted 10 fold with ChIP dilution buffer (16.7 mM Tris-HCl [pH 7.9], 1.2 mM EDTA, 167 mM NaCl, 1.1% Triton X-100 and 0.01% SDS) and then pre-cleared with Protein A Sepharose 4 Fast Flow (GE Healthcare). An antibody was added to the pre-cleared sample solution and incubated overnight at 4°C. Antibody-protein-DNA complexes were incubated with Protein A Sepharose at 4°C for 1 hr and then the beads were washed with Low Salt Wash Buffer (20 mM Tris-HCl [pH 7.9], 2 mM EDTA, 150 mM NaCl, 1% Triton X-100 and 0.1% SDS), High Salt Wash Buffer (20 mM Tris-HCl [pH 7.9], 2 mM EDTA, 500 mM NaCl, 1% Triton X-100 and 0.1% SDS), LiCl Wash Buffer (10 mM Tris-HCl [pH 7.9], 1 mM EDTA, 0.25 M LiCl, 1% NP-40 and 1% deoxycholic acid) and TE (10 mM Tris-HCl [pH 7.9] and 1 mM EDTA) successively. Protein-DNA complexes were eluted from the beads with elution buffer (1% SDS, 100 mM NaHCO3 and 10 mM DTT) and crosslinking was reversed by incubation at 65°C for 4 hr. After treatment with Proteinase K, DNAs were recovered by phenol/chloroform extraction and ethanol precipitation. Obtained DNA samples were subjected to qPCR as described above.
Table 2 shows primer sequences for 5′ inverted terminal region (5′ ITR), the E1A enhancer (E1A enh), the E1A promoter (E1A pro), the E1A coding region (E1A cds), the E1A 3′ region (E1A 3′), the E1B promoter (E1B pro), the IX promoter (IX pro), the ML promoter (MLP), the VA I gene (VA I), the Hexon ORF (Hexon), the E2B ORF (E2B orf), the E2 late promoter (E2late pro), the E3 promoter (E3 pro), the E4 ORF3 (E4 orf) and the E4 promoter regions (E4 pro).
siRNA-mediated knockdown, quantification of viral DNA and western blot assays
These experiments were carried out essentially as described previously12,14,19. siRNA targeted for CTCF was commercially purchased (Stealth siRNA; Invitrogen). siRNAs were introduced into cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Total DNAs were purified by treatment with Proteinase K, followed by phenol/chloroform extraction and ethanol precipitation, as described previously19. Quantitative determination of viral DNA was carried out by qPCR as described above. For western blot analyses, cell lysates were subjected to SDS-PAGE and proteins were transferred to a PVDF membrane (Millipore). The membrane was incubated with primary antibodies, followed by incubation with secondary antibodies conjugated with horseradish peroxidase (GE Healthcare). The blot was developed using Chemi-Lumi One (Nacalai tesque).
References
Raab, J. R. & Kamakaka, R. T. Insulators and promoters: closer than we think. Nat Rev Genet 11, 439–46 (2010).
Phillips, J. E. & Corces, V. G. CTCF: master weaver of the genome. Cell 137, 1194–211 (2009).
Kim, T. H. et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–45 (2007).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–37 (2007).
Stedman, W. et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27, 654–66 (2008).
Wendt, K. S. et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 (2008).
Yusufzai, T. M., Tagami, H., Nakatani, Y. & Felsenfeld, G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13, 291–8 (2004).
Kang, H., Wiedmer, A., Yuan, Y., Robertson, E. & Lieberman, P. M. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog 7, e1002140 (2011).
Tempera, I., Klichinsky, M. & Lieberman, P. M. EBV latency types adopt alternative chromatin conformations. PLoS Pathog 7, e1002180 (2011).
Giberson, A. N., Davidson, A. R. & Parks, R. J. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res 40, 2369–76 (2012).
Haruki, H., Okuwaki, M., Miyagishi, M., Taira, K. & Nagata, K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J Virol 80, 794–801 (2006).
Komatsu, T., Haruki, H. & Nagata, K. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res 39, 889–901 (2011).
Thomas, G. P. & Mathews, M. B. DNA replication and the early to late transition in adenovirus infection. Cell 22, 523–33 (1980).
Komatsu, T. & Nagata, K. Replication-uncoupled histone deposition during adenovirus DNA replication. J Virol 86, 6701–11 (2012).
Swaminathan, S. & Thimmapaya, B. Regulation of adenovirus E2 transcription unit. Curr Top Microbiol Immunol 199, 177–94 (1995).
Matsui, T., Murayama, M. & Mita, T. Adenovirus 2 peptide IX gene is expressed only on replicated DNA molecules. Mol Cell Biol 6, 4149–54 (1986).
Maxfield, L. F. & Spector, D. J. Readthrough activation of early adenovirus E1b gene transcription. J Virol 71, 8321–9 (1997).
Fisher, D. & Méchali, M. Vertebrate HoxB gene expression requires DNA replication. EMBO J 22, 3737–48 (2003).
Samad, M. A., Komatsu, T., Okuwaki, M. & Nagata, K. B23/nucleophosmin is involved in regulation of adenovirus chromatin structure at late infection stages, but not in virus replication and transcription. J Gen Virol 93, 1328–38 (2012).
Acknowledgements
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.N.).
Author information
Authors and Affiliations
Contributions
T.K. and K.N. designed the study; T.K. and T.S. performed the experiments; T.K. and K.N. prepared the manuscript; All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Info File #1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Komatsu, T., Sekiya, T. & Nagata, K. DNA replication-dependent binding of CTCF plays a critical role in adenovirus genome functions. Sci Rep 3, 2187 (2013). https://doi.org/10.1038/srep02187
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02187
This article is cited by
-
CTCF interacts with the lytic HSV-1 genome to promote viral transcription
Scientific Reports (2017)
-
Polycomb repressive complex 2 facilitates the nuclear export of the influenza viral genome through the interaction with M1
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.