Abstract
New small molecules that regulate the step-wise differentiation of human pluripotent stem cells into dopaminergic neurons have been identified. The steroid, guggulsterone, was found to be the most effective inducer of neural stem cells into dopaminergic neurons. These neurons are extensively characterized and shown to be functional. We believe this new approach offers a practical route to creating neurons of sufficient quality to be used to treat Parkinson's disease patients.
Similar content being viewed by others
Introduction
Parkinson's disease (PD) is a neurodegenerative disorder caused by a progressive degeneration of midbrain dopamine (DA) neurons in the substantia nigra pars compacta. The degeneration of DA neurons causes a gradual dysfunction of the motor system leading to symptoms such as tremor, rigidity and bradykinesia, among others. There is currently no cure for PD and although treatments such as deep brain stimulation and levodopa can alleviate some of the symptoms, they tend to lose efficacy over time. However, the localized nature of the loss of DA neurons in the substantia nigra (SN) makes cell replacement therapy an attractive approach to treating Parkinson's disease (PD) patients. Implantation of neuronal cells such as neural stem cells (NSCs) and DA neurons have already been shown to improve the motor symptoms in PD animal models1,2,3,4,5. In order for stem cell based therapies for PD to become a reality it is critical to be able to generate a homogeneous population of NSCs which will in turn generate functional DA neurons either in situ or in vitro, depending on whether the terminal differentiation occurs in the patient's brain or in culture. In this communication we report the derivation of a high-purity population of DA neurons from human pluripotent stem cells (hPSCs) in a robust and reproducible manner via a stable NSC stage, where the cells can be expanded and cryopreserved, using newly developed chemically directed differentiation methods.
Results
Derivation of NSCs from hPSCs with under defined chemical conditions
To derive NSCs, we identified two potent neural inducers, SB218078 and DMH-1 using a high-throughput screening strategy ( Fig. 1a ). SB218078 is a structural homolog of stauprimide, which has been shown to prime embryonic stem cells for differentiation6 and DMH-1 is a homolog of dorsomorphin, which efficiently induces neural conversion in hPSCs7. To derive NSCs, primitive neuroepithelium was induced by switching the hPSCs grown on Matrigel from StemPro to N2B27 medium (see Methods) supplemented with 5 μM SB218078 and 1 μM DMH-1. After 6 days in the presence of these neural inducers, Pax6, an early marker of neural induction, was significantly up-regulated ( Fig. 1b ). On day 7, the neuralized hPSCs were switched to NSC medium (see Methods) and then dissociated to derive proliferative NSCs. After 4 passages, the hPSC derived NSC (hPSC-NSC) population was 98% positive for Nestin, 96% for Musashi-1, 95% for PAX6, 0% for OCT4 ( Fig. 1c and Fig. 1d ). Gene expression microarray analysis revealed up-regulation of putative NSC markers including FABP7, BRN2, SOX3, ST6GALNAC5, CXCR4, DCX, NES and MSI ( Fig. 1e ). FABP7 encodes for a brain fatty acid binding protein involved in CNS development and is highly expressed in NSCs8. ST6GALNAC5 encodes for alpha 2,6-sialyltransferase, which mediates the passage of cells through the blood brain barrier9. Real-time PCR (RT-PCR) data further confirmed the up-regulation of NSC markers and, most importantly, revealed the complete down-regulation of the pluripotency markers OCT-4 and NANOG ( Fig. 1f ), leading to the conclusion that a highly enriched population of neural stem cells had been obtained. These NSCs are suitable for further expansion, cryopreservation and differentiation, making them a practical source for DA neurons.
Generation of hPSC derived NSCs through small molecules.
(a) Diagram showing small molecule screening strategy for the neural induction of hPSCs. (b) hPSCs treated with SB218078 and DMH1 for seven days and stained for PAX6. Scale bar is 100 μm. (c) Expanded hPSC-NSCs stained for Nestin, Musashi and PAX6. Scale bar is 100 μm. (d) FACS analyses of hPSC-NSC population stained for Musashi, Nestin, PAX6 and OCT4. (e) Gene expression microarray analysis showing fold expression induction of neural markers from hPSCs to NSCs; n = 2. (f) Gene expression analysis by RT-PCR of hPSC and hPSC-NSCs. Mean ± s.e.m., two-tailed Student's t-test: n = 3–5; α = 0.05; **P < 0.01; *P < 0.05.
High-throughput screening assay for DA neuron differentiation
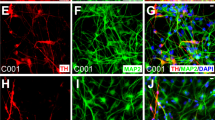
To obtain DA neurons, hPSC-NSCs were first primed into DA precursors with 100 ng/mL FGF8 and 2 μM purmorphamine for 7 days and then used to screen for small molecules that induce terminal dopaminergic differentiation ( Fig. 2a ). The DA precursors were plated at 2000 cells/well of a 96-well plate and treated with a small molecule library (Tocris 1120 biologically active compounds) at 2.5 μM for two weeks ( Fig. 2a ). Dopaminergic differentiation was assessed by neurite outgrowth and ELISA analysis for dopamine release ( Fig. 2a ). From this screen, guggulsterone (GS), a naturally occurring steroid10, was identified among the best inducers of dopaminergic differentiation ( Fig. 2a ). After 30 days of differentiation, GS-treated neurons appeared mature with elaborate neurite extensions. At this stage, the derived cells expressed not only the major neuronal marker β-III-tubulin (TUJ1), but also the important DA neuron markers tyrosine hydroxylase (TH), the dopamine transporter (Dat), Foxa2, Nurr-1 and Girk2 ( Fig. 2b ). Furthermore, FACS analysis revealed that more than 97% of the cells gated were TH positive ( Fig. 2c ). Gene expression microarray analysis indicated up-regulation of dopaminergic and neuronal associated markers such as MAPT, FOXA2, SYT4, FOXA1, DDC, ASCL1 and PINK1 ( Fig. 2d ). SYT4 (Synaptotagmin 4) plays an important role in synaptic plasticity and dopamine release11. ASCL1 is expressed in the ventral mesencephalon12 and indicates the derived cells are of the appropriate phenotype. Further analysis of the gene expression of hPSC, NSC, DA neurons and tissue from the substantia nigra revealed that the hPSC-derived DA neurons have acquired elements of the gene expression signature characteristic of cells undergoing neural differentiation toward cells present in the SN and that they are developmentally located after the NSC and before the SN stage ( Supplementary Fig. S1 ). RT-PCR further confirmed the DA neuron identity with the characteristic expression of markers such as PAX5, LMX1B, PITX3, NURR1, LMX1A, EN1, GIRK2, DDC and VMAT2 ( Fig. 2e ). Most importantly, compared to control cells (NSCs treated for 1 week with FGF8 and purmorphamine and 2 weeks with 0.1% DMSO instead of GS), GS-treated cells demonstrated a five-fold increase in dopamine secretion as determined by ELISA ( Fig. 2f ). Whole cell patch-clamp electrophysiology ( Fig. 2g , Fig. 2h and Fig. 2i ) revealed that after 90 days of GS treatment, the DA neurons displayed inward Na+ current in voltage-clamp mode ( Fig. 2h ) and fired action potentials when current was injected in the current-clamp mode ( Fig. 2i ). Spontaneous action potentials were recorded in whole cultures of GS-treated neurons using an Axion Biosystems Muse microelectrode array (MEA) system ( Fig. 2j ). Approximately one third of the 64 total electrodes in MEA dishes showed moderate activity (>5 spikes/min) ( Supplemental Fig. S2, Supplementary Video S1 ). These neurons are characterized by two basic firing phenotypes: those that fire periodic bursts of spikes and those that regularly fire single spikes ( Fig. 2j and Fig. 2k ).
Guggulsterone promotes generation of DA neurons.
(a) Diagram showing small molecule screening strategy for the differentiation of hPSC derived NSCs (hPSC-NSC) into DA neurons. (b) Staining for dopaminergic markers after 30 days of guggulsterone (GS) treatment. Scale bar is 100 μm. (c) FACS analysis of derived DA neurons stained for TH. (d) Fold induction of dopaminergic markers from NSCs to DA neurons as determined by gene expression microarray analysis; n = 2 (e) Relative gene expression by RT-PCR in DA neurons, NSCs and undifferentiated hPSCs. Mean ± s.e.m., one-factor ANOVA with Dunnett test comparing expression of DA neurons and NSCs against hPSC controls: n = 3–5; α = 0.05; ***P < 0.001; **P < 0.01; *P < 0.05. (f) Dopamine release assay of GS-treated neurons versus control cells (NSCs treated for 1 week with 100 ng/ml FGF8 and 2 μM purmorphamine and 2 weeks with 0.1% DMSO instead of GS). Mean ± s.e.m., two-tailed Student's t-test: n = 3; α = 0.05; **P < 0.01. (g) Representative phase-contrast image of a whole-cell patch-clamped GS-treated neuron. Scale bar is 10 μm. (h) Sodium current elicited with application of voltage ramp from −60 to +50 mV to cell in (g) in voltage clamp mode. Ramp parameters and waveform are shown in red superimposed on the current trace. The blue arrow indicates the point at which the stimulation ramp crossed the 0 mV value. (i) Action potential elicited from cell in (g) by injection of 500 pA for 200 ms in current clamp mode. Action potentials were found in 7 out of 20 cells patch-clamped. (j) Extracellular microelectrode array (MEA) recordings of GS-treated neurons, showing 10 s voltage traces for three representative electrodes. A single spike for each electrode is expanded at right. (k) Interspike interval histograms for electrodes in (j) showing: (↓↓) peaks associated with intervals between spikes within a burst and; (↓) those for longer intervals between bursts, or between individual spikes on non-bursting electrodes. Similar results were obtained with 2 MEA dishes.
Discussion
Obtaining a high purity population of neuronal cells is critical for the development of cell therapies for Parkinson's disease, disease modeling and high-throughput drug discovery. The use of small molecules to direct differentiation of pluripotent stem cells toward a specific linage represents a powerful approach to generate particular cell types with potential clinical applications13. To date, one of the best methods for the production of human dopaminergic neurons from hES cells is via a novel floor plate-based (FP) strategy1. The stepwise method outlined here is straightforward, efficiently promoting the differentiation to DA neurons with only one small molecule guggulsterone (GS) used in the final stage compared to a more complex mixture of growth factors and chemicals, including BDNF, GDNF, TGFβ3, DAPT, dbcAMP and ascorbic acid used in aforementioned FP method. In fact, we found that DA neurons derived using GS, as a substitute for the aforementioned biologic and chemical signaling factors, secrete three-fold higher dopamine levels in vitro (Supplementary Figure S3). Additionally the GS-derived neurons have gene and protein expression profiles typical for DA neurons as well as in vitro functionality demonstrated by whole-cell patch clamp and MEA recordings. This observed robust pacemaking is phenotypical for DA neurons from the substantia nigra1,14.
In summary, the results reported here demonstrate that the bioactive small molecule GS can be used to dramatically enhance neuronal differentiation towards dopaminergic neurons. This newly reported stepwise method allows the establishment of a scalable, feeder- and xeno- free protocol, to produce pure populations of clinical-grade neuronal cells from hPSCs and is an important advancement towards developing an effective treatment for Parkinson's disease.
Methods
Culture of hPSCs
The hPSC lines [human embryonic stem cell line WA-09 and human parthenogenetic stem cell lines LLC2P and LLC12PH (International Stem Cell Corporation)15,16] were first maintained on a mitomycin-C inactivated mouse embryonic fibroblast (Millipore) feeder layer in embryonic stem cell medium: Knock Out DMEM/F12 (Life Technologies), 2 mM L-glutamine (GlutaMax-I, Invitrogen), 0.1 mM MEM nonessential amino acids (Life Technology), 0.1 mM β-mercaptoethanol (Life Technologies), penicillin/streptomycin/amphotericin B (100 U/100 μg/250 ng) (MP Biomedicals) and 5 ng/ml bFGF (Peprotech). Cells were passaged with dispase (Life Technologies) every 5–7 days with split ratio of 1:4 or 1:6. The hPSCs were then transferred to Matrigel (BD Biosciences) coated plates and grown with Stem Pro hESC SFM medium (Invitrogen): DMEM/F12 with GlutaMAX, 1× STEMPRO hESC SFM Growth Supplement, 1.8% Bovine Serum Albumin, 8 ng/mL bFGF and 0.1 mM 2-Mercaptoethanol.
Derivation and growth of hPSC-NSCs
hPSC-NSCs were derived by treating undifferentiated hPSCs growing under feeder-free culture conditions with a chemical combination consisting of SB218078 (5 μM) plus DMH-1 (1 μM) in N2B27 Medium [Knockout DMEM/F12, 1× GlutaMax, 1× N2/B27 Supplement (Invitrogen)] for 7 days. The neuralized hPSCs were then dissociated with Accutase (Sigma) and grown on Matrigel coated plates in NSC medium [KnockOut DMEM/F12, 2% StemPro Neural Supplement, 1× GlutaMAX, 20 ng/ml bFGF and 20 ng/ml EGF] for ≥ 4 passages to generate and highly pure and homogeneous population of hpSC-NSCs.
High-throughput screening assay for DA neuron differentiation
hPSC-NSCs were treated with purmorphamine (2 μM) and FGF8 (100 ng/mL) in NB medium [NeuroBasal medium, 1× GlutaMAX, 1× N2/B27 Supplement (Invitrogen)] for 7 days. The primed NSCs were then dissociated with Accutase (Sigma) and seeded at a concentration of 20,000 cells/mL into Matrigel- coated 96 well plates and then treated with small molecule library (Torcris 1120 Biologically Active Compounds) at 2.5 μM (final concentration) or control (0.1% DMSO) for 2 weeks. Two weeks after treatment with small molecules, all wells were visually observed and scored based on neurite density and dopamine release. For measuring neurite density, cells were fixed with 4% paraformaldehyde and phase contrast images were acquired using Cellavista Cell Imaging System (Roche Applied Science) from randomly selected fields. Neurite density was measured using the Cellavista density software image processing program. Each experimental condition was done in duplicate wells and at least three independent experiments were conducted to acquire the final results.
Dopamine ELISA
Conditioned medium was collected from samples containing 50,000 cells per well and centrifuged at 3000 RPM to remove cell debris. Supernatant was then collected and analyzed for quantitative determination of dopamine using a commercial dopamine ELISA assay kit (Cusabio). Data analysis was performed using the microtiter plate reader (BIO-TEK Synergy 2). The results presented are from three independent experiments (n = 3) and represented as mean ± standard error of mean (s.e.m.) and statistical analysis was performed using the two-tailed Student's t-test with a confidence level of 95% (α = 0.05) with statistical significance of P < 0.05.
Whole-cell patch clamp electrophysiology
For electrophysiological recordings, the cells were transferred to a recording chamber mounted on an inverted Olympus microscope. During the recordings the temperature was maintained at 31–32°C and the cells perfused at constant flow of 1 ml/min. with 150 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 10 mM Glucose at pH 7.30 and 315 mOsm. Whole-cell recordings were obtained using borosilicate electrodes with tip resistance of 2–3 MΩ, filled with 130 mM K-Gluconate, 5 mM NaCl, 5 mM KCl, 10 mM HEPES, 20 mM Sucrose, 0.3 mM EGTA, 3 mM MgATP at pH 7.30 and 290 mOsm. Data were acquired using an HEKA EPC-10 digitizer/amplifier. Once whole-cell access was achieved the resting membrane potential was immediately recorded; the cells were then allowed to equilibrate for 3–5 min. From a resting membrane potential of −70 mV, the cells were then depolarized in current-clamp mode with 200 ms current injections, delivered at 0.2 Hz. and with increasing amplitude from 50 pA to up to 1000 pA to induce action potentials. The presence of inward or outward currents was determined by applying a voltage ramp. Cells with unstable recording configuration or for which the series resistance became larger than 15 MΩ were excluded from the study. We recorded 20 different cells and obtained action potentials from 7 patch-clamped cells.
Microarray analysis
Total RNA was extracted from collected from duplicate sample pellets (RNeasy; Qiagen) according to the manufacturer's protocol. RNA quantity (Qubit RNA BR Assay Kits; Invitrogen) and quality (RNA6000 Nano Kit; Agilent) was determined to be optimal for each sample before further processing. 200 ng RNA per sample was amplified using the Illumina Total Prep RNA Amplification Kit according to manufacturer's protocol and quantified as above. 750 ng biotinylated RNA per sample was hybridized to Illumina HT-12v4 Expression BeadChips, scanned with an Illumina iScan Bead Array Scanner and quality controlled in GenomeStudio and the lumi bioconductor package. All RNA processing and microarray hybridizations were performed according to manufacturer's protocols. In GenomeStudio, probes were filtered for those detected with a P value of 0.01 in at least one sample and exported for normalization in R. Raw probe expression values were transformed and normalized using the robust spline normalization (RSN) as implemented in the lumi R/Bioconductor package. To obtain the Venn Diagram, Qlucore Omni Explorer was used. Transcripts that were differentially expressed between pairs of cell types (hPSC vs Substantia Nigra, hPSC vs NSC and hPSC vs Dopaminergic neurons) were identified using two-tailed Student's t-test with P-value cut-off of <0.05 and a variance cut-off of >0.005. The sets of differentially expressed probes were then compared to each other. The gene expression array data is available at the NCBI GEO database under the accession designation GSE42265.
Statistical analysis
The results are presented as mean ± standard error of mean (s.e.m.) and statistical analysis was performed using a confidence level of 95% (α = 0.05) with two-tailed Student's t-test for comparing two groups or one factor ANOVA with Dunnett test for comparing multiple groups against control. The criterion for statistical significance for all tests was P < 0.05.
Additional methods
Immunocytochemistry, flow cytometry and RT-PCR have been previously described17 and details for these experimental procedures as well as for Microelectrode Array (MEA) System are given in the Supplementary Information.
References
Kriks, S. et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 480, 547–551 (2011).
Hargus, G. et al. Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. Proc Natl Acad Sci USA 107, 15921–15926 (2010).
Ganat, Y. M. et al. Identification of embryonic stem cell-derived midbrain dopaminergic neurons for engraftment. J Clin Invest. 122, 2928–2939 (2012).
Parish, C. L. et al. Wnt5a-treated midbrain neural stem cells improve dopamine cell replacement therapy in parkinsonian mice. J Clin Invest. 118, 149–160 (2008).
Redmond, D. E. Jr. et al. Behavioral improvement in a primate Parkinson's model is associated with multiple homeostatic effects of human neural stem cells. Proc Natl Acad Sci USA 104, 12175–12180 (2007).
Zhu, S. et al. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell 4, 416–426 (2009).
Zhou, J. et al. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells 28, 1741–1750 (2010).
Yun, S. W. et al. Neural stem cell specific fluorescent chemical probe binding to FABP7. Proc Natl Acad Sci USA 109, 10214–10217 (2012).
Bos, P. D. et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009).
Urizar, N. L. et al. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 296, 1703–1706 (2002).
Mendez, J. A., Bourque, M. J., Fasano, C., Kortleven, C. & Trudeau, L. E. Somatodendritic dopamine release requires synaptotagmin 4 and 7 and the participation of voltage-gated calcium channels. J Biol Chem. 286, 23928–23937 (2011).
Nelander, J., Hebsgaard, J. B. & Parmar, M. Organization of the human embryonic ventral mesencephalon. Gene Expr Patterns 9, 555–561 (2009).
Surmacz, B., Fox, H., Gutteridge, A., Lubitz, S. & Whiting, P. Directing differentiation of human embryonic stem cells toward anterior neural ectoderm using small molecules. Stem Cells 30, 1875–1884 (2012).
Guzman, J. N., Sanchez-Padilla, J., Chan, C. S. & Surmeier, D. J. Robust pacemaking in substantia nigra dopaminergic neurons. J Neurosci. 29, 11011–11019 (2009).
Revazova, E. S. et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells 9, 432–449 (2007).
Revazova, E. S. et al. HLA homozygous stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells 10, 11–24 (2008).
Isaev, D. A. et al. In vitro differentiation of human parthenogenetic stem cells into neural lineages. Regen Med. 7, 37–45 (2012).
Acknowledgements
We thank Prof. Loring from the Scripps Research Institute for valuable scientific advice. LCL was supported by NIH K12 HD001259 and The Hartwell Foundation. FSB was supported by The Hartwell Foundation. This work was partially supported to CIRM TR-01250 to Prof. Loring.
Author information
Authors and Affiliations
Contributions
R.G., I.G., T.A., G.W., A.O., A.N., F.B., L.L., M.B. performed experiments. R.G., I.G., R.S. designed the study. R.S., I.G., R.G., J.C. wrote the manuscript. E.S. provided scientific advice.
Ethics declarations
Competing interests
R.G., I.G., A.O., T.A., G.W., A.N., J.C.; R.S. are employees and stockholders of International Stem Cell Corporation. M.B. is an employee and stockholder of Axion Biosystems. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Video S1.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Gonzalez, R., Garitaonandia, I., Abramihina, T. et al. Deriving dopaminergic neurons for clinical use. A practical approach. Sci Rep 3, 1463 (2013). https://doi.org/10.1038/srep01463
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01463
This article is cited by
-
3D bioprinting patient-derived induced pluripotent stem cell models of Alzheimer’s disease using a smart bioink
Bioelectronic Medicine (2023)
-
Challenges in the clinical advancement of cell therapies for Parkinson’s disease
Nature Biomedical Engineering (2023)
-
Restoring Function to Dopaminergic Neurons: Progress in the Development of Cell-Based Therapies for Parkinson’s Disease
CNS Drugs (2020)
-
Neural Stem Cell Tumorigenicity and Biodistribution Assessment for Phase I Clinical Trial in Parkinson’s Disease
Scientific Reports (2016)
-
On the road to bioartificial organs
Pflügers Archiv - European Journal of Physiology (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.