Key Points
-
Motility is one of the most dynamic features of the microbial world. The ability to swim in liquid or crawl on surfaces frequently governs how microorganisms interact with their physical and chemical environment, and underpins a myriad of microbial processes.
-
The ability to resolve temporal dynamics through time-lapse imaging and the precise control of the physicochemical microenvironment afforded by microfluidics offer powerful new opportunities to study the motility adaptations of microorganisms and thereby further our understanding of their ecology.
-
Dynamic microscale imaging has shown how individual swimming microorganisms disturb the fluid in their surroundings, and how this hydrodynamic signature affects their motility near surfaces as well as in dense-cell suspensions. The same technique has revealed new motility adaptations of microorganisms, in particular the flicking behaviour used by many marine bacteria to turn.
-
Tracking swimming microorganisms in precisely controlled chemical gradients created using microfluidic devices has revealed that microorganisms are capable of refined rescaling responses in their chemotactic behaviour, which ensure high performance under a wide range of environmental conditions. In the environment, and in particular in the ocean, the strong chemotactic responses of microorganisms can be important in determining associations with larger organisms, consuming dissolved organic matter and ultimately affecting biogeochemistry.
-
An important but often neglected set of microbial interactions are those between cells and their physical environment — chiefly, surfaces and fluid flow. Recent imaging-based microfluidic studies have revealed that hydrodynamic and surface-induced forces can strongly bias the direction of migration of microorganisms. These forces, for example, induce upstream swimming or preferential cell accumulations in regions of high-velocity gradients, affecting the transport of microorganisms and the colonization of surfaces that leads to biofilm formation.
Abstract
Motility is one of the most dynamic features of the microbial world. The ability to swim or crawl frequently governs how microorganisms interact with their physical and chemical environments, and underpins a myriad of microbial processes. The ability to resolve temporal dynamics through time-lapse video microscopy and the precise control of the physicochemical microenvironment afforded by microfluidics offer powerful new opportunities to study the many motility adaptations of microorganisms and thereby further our understanding of their ecology. In this Review, we outline recent insights into the motility strategies of microorganisms brought about by these techniques, including the hydrodynamic signature of microorganisms, their locomotion mechanics, chemotaxis, their motility near and on surfaces, swimming in moving fluids and motility in dense microbial suspensions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Xie, L., Altindal, T., Chattopadhyay, S. & Wu, X.-L. Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc. Natl Acad. Sci. USA 108, 2246–2251 (2011). This study reported the discovery of the 'flick', a new reorientation mechanism found among marine bacteria, which makes their motility drastically different from the run-and-tumble motility observed in E. coli.
Son, K., Guasto, J. S. & Stocker, R. Bacteria can exploit a flagellar buckling instability to change direction. Nat. Phys. 9, 494–498 (2013).
Zhao, K. et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497, 388–391 (2013). This study mapped the chemical trails of individual bacteria on a surface, demonstrating that matrix-rich regions are self-reinforcing and form the skeleton of biofilms.
Wang, P. et al. Robust growth of Escherichia coli. Curr. Biol. 20, 1099–1103 (2010).
Zhang, Q. et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333, 1764–1767 (2011).
Jin, F., Conrad, J. C., Gibiansky, M. L. & Wong, G. C. L. Bacteria use type-IV pili to slingshot on surfaces. Proc. Natl Acad. Sci. USA 108, 12617–12622 (2011). This study revealed that P. aeruginosa twitching on surfaces are capable of a rapid slingshot motion that can efficiently reorient cells.
Gibiansky, M. L. et al. Bacteria use type IV pili to walk upright and detach from surfaces. Science 330, 197 (2010).
Persat, A., Stone, H. A. & Gitai, Z. The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat. Commun. 5, 3824 (2014).
Hol, F. J. H. & Dekker, C. Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346, 1251821 (2014).
Sackmann, E. K., Fulton, A. L. & Beebe, D. J. The present and future role of microfluidics in biomedical research. Nature 507, 181–189 (2014).
Rusconi, R., Garren, M. & Stocker, R. Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 43, 65–91 (2014).
Wessel, A. K., Hmelo, L., Parsek, M. R. & Whiteley, M. Going local: technologies for exploring bacterial microenvironments. Nat. Rev. Microbiol. 11, 337–348 (2013).
Guasto, J. S., Rusconi, R. & Stocker, R. Fluid mechanics of planktonic microorganisms. Annu. Rev. Fluid Mech. 44, 373–400 (2012).
Elgeti, J., Winkler, R. G. & Gompper, G. Physics of microswimmers-single particle motion and collective behavior: a review. Rep. Prog. Phys. 78, 056601 (2015).
Drescher, K., Dunkel, J., Cisneros, L. H., Ganguly, S. & Goldstein, R. E. Fluid dynamics and noise in bacterial cell–cell and cell–surface scattering. Proc. Natl Acad. Sci. USA 108, 10940–10945 (2011). This study reported the first experimental quantification of the flow field around a single swimming E. coli bacterium.
Lighthill, M. J. Mathematical Biofluiddynamics (Society for Industrial and Applied Mathematics, 1975).
Wu, X. L. & Liebchaber, A. Particle diffusion in a quasi-two-dimensional bacterial bath. Phys. Rev. Lett. 84, 3017–3020 (2010).
Zhang, H.-P., Be'er, A., Florin, E.-L. & Swinney, H. L. Collective motion and density fluctuations in bacterial colonies. Proc. Natl Acad. Sci. USA 107, 13626–13630 (2010).
Dombrowski, C., Cisneros, L., Chatkaew, S., Goldstein, R. E. & Kessler, J. O. Self-concentration and large-scale coherence in bacterial dynamics. Phys. Rev. Lett. 93, 2–5 (2004).
Berke, A. P., Turner, L., Berg, H. C. & Lauga, E. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys. Rev. Lett. 101, 038102 (2008).
Berg, H. C. E. coli in Motion (Springer, 2004).
Turner, L., Ryu, W. S. & Berg, H. C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182, 2793–2801 (2000).
Stocker, R. Reverse and flick: hybrid locomotion in bacteria. Proc. Natl Acad. Sci. USA 108, 2635–2636 (2011).
Leifson, E., Cosenza, B. J., Murchelano, R. & Cleverdon, R. C. Motile marine bacteria I. techniques, ecology, and general characteristics. J. Bacteriol. 87, 652–666 (1964).
Xie, L. & Wu, X. L. Bacterial motility patterns reveal importance of exploitation over exploration in marine microhabitats. part I: theory. Biophys. J. 107, 1712–1720 (2014).
Taktikos, J., Stark, H. & Zaburdaev, V. How the motility pattern of bacteria affects their dispersal and chemotaxis. PLoS ONE 8, e81936 (2014).
Wadhams, G. H. & Armitage, J. P. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 5, 1024–1037 (2004).
Tu, Y. H. Quantitative modeling of bacterial chemotaxis: Signal amplification and accurate adaptation. Annu. Rev. Biophys. 42, 337–359 (2013).
Stocker, R. & Seymour, J. R. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76, 792–812 (2012).
Stocker, R. Marine microbes see a sea of gradients. Science 338, 628–633 (2012).
Ahmed, T., Shimizu, T. S. & Stocker, R. Microfluidics for bacterial chemotaxis. Integr. Biol. 2, 604–629 (2010).
Kalinin, Y. V., Jiang, L., Tu, Y. & Wu, M. Logarithmic sensing in Escherichia coli bacterial chemotaxis. Biophys. J. 96, 2439–2448 (2009).
Fechner, G. T., Adler, H. E., Howes, D. H. & Boring, E. G. Elementary Psychophysics (Holt, 1966).
Rieke, F. & Rudd, M. E. The challenges natural images pose for visual adaptation. Neuron 64, 605–616 (2009).
Lazova, M. D., Ahmed, T., Bellomo, D., Stocker, R. & Shimizu, T. S. Response rescaling in bacterial chemotaxis. Proc. Natl Acad. Sci. USA 108, 13870–13875 (2011). This study revealed experimentally that E. coli is capable of rescaling its chemotactic response, a process termed fold-change detection, which ensures high chemotactic sensitivity across a broad range of environmental conditions.
Zhu, X. et al. Frequency-dependent Escherichia coli chemotaxis behavior. Phys. Rev. Lett. 108, 128101 (2012).
Kalinin, Y., Neumann, S., Sourjik, V. & Wu, M. Responses of Escherichia coli bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio. J. Bacteriol. 192, 1796–1800 (2010). This study was the first to use microfluidics to examine the chemotactic decision-making process of E. coli cells that were exposed to two simultaneous chemical gradients.
Blackburn, N. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science 282, 2254–2256 (1998).
Bell, W. & Mitchell, R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143, 265–277 (1972).
Barbara, G. M. & Mitchell, J. G. Bacterial tracking of motile algae. FEMS Microbiol. Ecol. 44, 79–87 (2003).
Stocker, R., Seymour, J. R., Samadani, A., Hunt, D. E. & Polz, M. F. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl Acad. Sci. USA 105, 4209–4214 (2008).
Seymour, J. R., Ahmed, T., Durham, W. M. & Stocker, R. Chemotactic response of marine bacteria to the extracellular products of Synechococcus and Prochlorococcus. Aquat. Microb. Ecol. 59, 161–168 (2010).
Seymour, J. R., Ahmed, T. & Stocker, R. Bacterial chemotaxis towards the extracellular products of the toxic phytoplankton Heterosigma akashiwo. J. Plank. Res. 31, 1557–1561 (2009).
Seymour, J. R., Simó, R., Ahmed, T. & Stocker, R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345 (2010).
Garren, M. et al. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007 (2014).
Seymour, J. R., Marcos & Stocker, R. Resource patch formation and exploitation throughout the marine microbial food web. Am. Nat. 173, E15–29 (2009).
Barbara, G. M. & Mitchell, J. G. Marine bacterial organisation around point-like sources of amino acids. FEMS Microbiol. Ecol. 43, 99–109 (2003).
Yawata, Y. et al. Competition–dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc. Natl Acad. Sci. USA 111, 5622–5627 (2014). This study revealed a competition–dispersal tradeoff among recently speciated sympatric marine bacteria, based on distinct behavioural interactions with particulate organic matter.
Persat, A. et al. The mechanical world of bacteria. Cell 161, 988–997 (2015).
O'Toole, G., Kaplan, H. B. & Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000).
Karimi, A., Karig, D., Kumar, A. & Ardekani, A. M. Interplay of physical mechanisms and biofilm processes: review of microfluidic methods. Lab. Chip 15, 23–42 (2015).
Watnick, P. & Kolter, R. Biofilm, city of microbes. J. Bacteriol. 182, 2675–2679 (2000).
Kearns, D. B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644 (2010).
Teschler, J. K. et al. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268 (2015).
Magariyama, Y. et al. Difference in bacterial motion between forward and backward swimming caused by the wall effect. Biophys. J. 88, 3648–3658 (2005).
Molaei, M., Barry, M., Stocker, R. & Sheng, J. Failed escape: solid surfaces prevent tumbling of Escherichia coli. Phys. Rev. Lett. 113, 68103 (2014).
Lauga, E., DiLuzio, W. R., Whitesides, G. M. & Stone, H. A. Swimming in circles: motion of bacteria near solid boundaries. Biophys. J. 90, 400–412 (2006). This study rationalized why many species of bacteria swim in circular trajectories when near a surface.
Utada, A. S. et al. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat. Commun. 5, 2913 (2014).
Conrad, J. C. et al. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys. J. 100, 1608–1616 (2011).
Rusconi, R. & Stocker, R. Microbes in flow. Curr. Opin. Microbiol. 25, 1–8 (2015).
Durham, W. M., Kessler, J. O. & Stocker, R. Disruption of vertical motility by shear triggers formation of thin phytoplankton layers. Science 323, 1067–1070 (2009).
Durham, W. M. et al. Turbulence drives microscale patches of motile phytoplankton. Nat. Commun. 4, 2148 (2013).
Rusconi, R., Guasto, J. S. & Stocker, R. Bacterial transport suppressed by fluid shear. Nat. Phys. 10, 212–217 (2014). This study revealed that the coupling of motility and flow can result in high levels of bacterial accumulation in certain regions of the flow, hampering chemotaxis and favouring surface attachment.
Hill, J., Kalkanci, O., McMurry, J. L. & Koser, H. Hydrodynamic surface interactions enable Escherichia coli to seek efficient routes to swim upstream. Phys. Rev. Lett. 98, 68101 (2007).
Kaya, T. & Koser, H. Direct upstream motility in Escherichia coli. Biophys. J. 102, 1514–1523 (2012).
Shen, Y., Siryaporn, A., Lecuyer, S., Gitai, Z. & Stone, H. A. Flow directs surface-attached bacteria to twitch upstream. Biophys. J. 103, 146–151 (2012).
Marcos, Fu, H. C., Powers, T. R. & Stocker, R. Bacterial rheotaxis. Proc. Natl Acad. Sci. USA 109, 4780–4785 (2012).
Meng, Y. Z. et al. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 187, 5560–5567 (2005). This study revealed that bacteria twitching on surfaces migrate upstream in the presence of fluid flow, owing to a hydrodynamic torque that orients them against the flow.
Bures, J. et al. Small intestinal bacterial overgrowth syndrome. World J. Gastroenterol. 16, 2978–2990 (2010).
Dunkel, J. et al. Fluid dynamics of bacterial turbulence. Phys. Rev. Lett. 110, 228102 (2013).
Sokolov, A., Goldstein, R., Feldchtein, F. & Aranson, I. Enhanced mixing and spatial instability in concentrated bacterial suspensions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 80, 1–8 (2009).
Saintillan, D. & Shelley, M. J. Active suspensions and their nonlinear models. Comptes Rendus Phys. 14, 497–517 (2013).
Kaiser, A. et al. Transport powered by bacterial turbulence. Phys. Rev. Lett. 112, 158101 (2014).
Butler, M. T., Wang, Q. & Harshey, R. M. Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl Acad. Sci. USA 107, 3776–3781 (2010).
Sitti, M. Miniature devices: voyage of the microrobots. Nature 458, 1121–1122 (2009).
Mesquita, A. R. & Hespanha, J. P. Jump control of probability densities with applications to autonomous vehicle motion. IEEE T. Automat. Contr. 57, 2588–2598 (2012).
Rubenstein, M., Cornejo, A. & Nagpal, R. Programmable self-assembly in a thousand-robot swarm. Science 345, 795–799 (2014).
Rusconi, R., Lecuyer, S., Guglielmini, L. & Stone, H. A. Laminar flow around corners triggers the formation of biofilm streamers. J. R. Soc. Interface 7, 1293–1299 (2010).
Drescher, K., Shen, Y., Bassler, B. L. & Stone, H. A. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl Acad. Sci. USA 110, 4345–4350 (2013).
Malfatti, F. & Azam, F. Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria. Aquat. Microb. Ecol. 58, 1–14 (2009).
Kiviet, D. J. et al. Stochasticity of metabolism and growth at the single-cell level. Nature 514, 376–379 (2014).
Rojas, E., Theriot, J. A. & Huang, K. C. Response of Escherichia coli growth rate to osmotic shock. Proc. Natl Acad. Sci. USA 111, 7807–7812 (2014).
Brumley, D. R., Wan, K. Y., Polin, M. & Goldstein, R. E. Flagellar synchronization through direct hydrodynamic interactions. eLife 3, e02750 (2014).
Berg, H. C. & Brown, D. A. Chemotaxis in Escherichia coli analyzed by three-dimensional tracking. Nature 239, 500–504 (1972).
Liu, B. et al. Helical motion of the cell body enhances Caulobacter crescentus motility. Proc. Natl Acad. Sci. USA 111, 11252–11256 (2014).
Wu, M. M., Roberts, J. W., Kim, S., Koch, D. L. & DeLisa, M. P. Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique. Appl. Environ. Microb. 72, 4987–4994 (2006).
Taute, K. M., Gude, S., Tans, S. J. & Shimizu, T. S. High-throughput 3D tracking of bacteria on a standard phase contrast microscope. Nat. Commun. 6, 8776 (2015).
Brumley, D. R., Polin, M., Pedley, T. J. & Goldstein, R. E. Metachronal waves in the flagellar beating of Volvox and their hydrodynamic origin. J. R. Soc. Interface. 12, 20141358 (2015).
Weibel, D. B., DiLuzio, W. R. & Whitesides, G. M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 5, 209–218 (2007).
Xia, Y. N. & Whitesides, G. M. Soft lithography. Annu. Rev. Mater. Sci. 28, 153–184 (1998).
Tabeling, P. Introduction to Microfluidics (Oxford Univ. Press, 2005).
Kantsler, V., Dunkel, J., Blayney, M., Goldstein, R. E. & Hyman, A. A. Rheotaxis facilitates upstream navigation of mammalian sperm cells. eLife 3, e02403 (2014).
Kim, S., Kim, H. J. & Jeon, N. L. Biological applications of microfluidic gradient devices. Integr. Biol. 2, 584–603 (2010).
Adler, M., Erickstad, M., Gutierrez, E. & Groisman, A. Studies of bacterial aerotaxis in a microfluidic device. Lab. Chip 12, 4835–4847 (2012).
Wong, I. & Ho, C. M. Surface molecular property modifications for poly(dimethylsiloxane) (PDMS) based microfluidic devices. Microfluid Nanofluid 7, 291–306 (2009).
Cheng, S. Y. et al. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab. Chip 7, 763–769 (2007).
Weibel, D. B. et al. Bacterial printing press that regenerates its ink: Contact-printing bacteria using hydrogel stamps. Langmuir 21, 6436–6442 (2005).
Lauga, E. & Powers, T. R. The hydrodynamics of swimming microorganisms. Rep. Prog. Phys. 72, 096601 (2009).
Acknowledgements
The authors gratefully acknowledge support through a Samsung Scholarship (to K.S.), a Human Frontier Science Program (HFSP) Cross-Disciplinary Fellowship (to D.R.B.) and a Marine Microbiology Initiative Investigator Award from the Gordon and Betty Moore Foundation (GBMF3783, to R.S.). The authors also thank G. Gorick for help with some of the figures.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (movie)
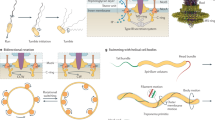
Motility mechanics. Many marine bacteria reorient by a 'flick', an off-axis deformation of the flagellum that enables bacteria with a single flagellum to change their direction of swimming. This video shows the flick process of Vibrio alginolyticus (see also Fig. 1c–f), recorded using high-speed, high-intensity dark-field microscopy (40X objective lens, 420 frames s−1). On the left is the raw video, on the right a processed version showing the (single, polar) flagellum in magenta. Note the buckling of the flagellum (see also Fig. 1e, 50–70 ms) shortly after the reversal in swimming direction ( Fig. 1e, 20 ms). This movie is reproduced from Ref. 2, Nature Publishing Group. (MOV 8234 kb)
Supplementary information S2 (movie)
Chemotaxis. Using chemotaxis, natural marine bacteria can cluster around photosynthetic diatoms, here Chaetoceros affinis, in response to the gradients in dissolved organic matter originating from the diatom (see also Fig. 2a). Courtesy of Steven Smriga and Vicente Fernandez, Department of Civil, Environmental and Geomatic Engineering, ETH Zurich, 8093 Zurich, Switzerland. (MOV 1950 kb)
Supplementary information S3 (movie)
Surface motility. Two-point tracking of a single Pseudomonas aeruginosa bacterium as it crawls along a surface (see also Fig. 3d). Markers 1 and 2 represent the leading and trailing poles, respectively. The video corresponds to 700 s in real time, with playback sped up by a factor of 40. This movie is reproduced with permission from Ref. 6, National Academy of Sciences. (AVI 6123 kb)
Supplementary information S4 (movie)
Motility in flow. Trajectory of a smooth-swimming Bacillus subtilis bacterium in a microfluidic channel (see also Fig. 4b). The raw video of the motile cell is shown first, followed by a replay in which the tracked cell trajectory (green) and position and orientation (red) are included. The flow in the channel is from left to right, and the video is recorded in the reference frame comoving with the mean speed of the flow (mean speed = 500 μm s−1, mean absolute shear rate = 2.5 s−1). The looped trajectory results from the velocity gradient generating a hydrodynamic torque that continually reorients the cell while it swims. The video was captured at 70.6 frames s−1 using dark-field microscopy, and is replayed 1.7 times slower than real time. This movie is reproduced from Ref. 63, Nature Publishing Group. (AVI 452 kb)
Glossary
- Soft lithography
-
A technique used for fabricating, at the micrometre to nanometre scale, features in elastomeric materials such as polydimethylsiloxane (PDMS).
- Defocused microscopy
-
A microscopic imaging technique whereby the distance of a microorganism ('into the plane') from the imaging plane is determined by matching its defocused ring size with a reference stack.
- Particle image velocimetry
-
(PIV). A method to measure the velocity field of a fluid based on the motion of many small passive tracer particles.
- Thermal fluctuations
-
A source of random noise in a system at equilibrium that induces diffusion of small particles.
- Rotational diffusion
-
For a swimming microorganism, this describes the continuous, random changes in swimming direction owing to thermal fluctuations (passive rotational diffusion) or to intrinsic imperfections (for example, wobbling) in the locomotion system (termed active rotational diffusion).
- Buckling
-
A sudden sideways failure of a structure subjected to compressive load.
- Logarithmic sensing
-
A sensing property in which cells respond to the relative gradient in a stimulus, ∇C/C, in which C is the magnitude and ∇C is the gradient magnitude of the stimulus.
- Förster resonance energy transfer
-
(FRET). A mechanism quantifying energy transfer between two light-sensitive molecules in which excitation is transferred from a donor molecule to an acceptor molecule without emission of a photon. In chemotactic transduction studies of Escherichia coli, FRET is used to measure the level of the chemotaxis signalling molecule phospho-CheY (CheYP) that controls flagellar reversals.
- Chemokinesis
-
The modulation of swimming speed in response to changes in the concentration of a chemical.
- Twitching
-
Crawling motion of bacteria on surfaces by means of pili.
- Digital holographic microscopy
-
A microscopic imaging technique where the position of an object 'into the plane' is encoded by the interference fringes it creates by diffracting light and can be reconstructed in post-processing to yield 3D information.
- Torque
-
The moment of the forces that act on an object, which quantifies their tendency to rotate the object.
- Mannose-sensitive haemagglutinin pili
-
(MSHA pili). One of three type IV pili, which play an important part in biofilm formation.
- Type IV pili
-
Thin, hair-like appendages present on the surface of many bacteria, involved in adherence to and motility on substrates.
- Jeffery orbit
-
Periodic rotational trajectory of an elongated particle (in this case, a microorganism) in a fluid velocity gradient, in which the angular speed varies with orientation relative to the flow.
- Laminar flow
-
Fluid motion devoid of turbulence and typically occurring as a smooth, orderly flow.
- Biofilm streamers
-
Conglomerates of cells and cell-secreted polymeric substances (exopolysaccharide) that are attached by one end to a surface and otherwise suspended in the flow. These biofilm structures exist in topographically complex environments exposed to fluid flow.
Rights and permissions
About this article
Cite this article
Son, K., Brumley, D. & Stocker, R. Live from under the lens: exploring microbial motility with dynamic imaging and microfluidics. Nat Rev Microbiol 13, 761–775 (2015). https://doi.org/10.1038/nrmicro3567
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3567
This article is cited by
-
Boosting microfluidic microbial fuel cells performance via investigating electron transfer mechanisms, metal-based electrodes, and magnetic field effect
Scientific Reports (2022)
-
Controlling pore-scale processes to tame subsurface biomineralization
Reviews in Environmental Science and Bio/Technology (2022)
-
Dynamic swimming pattern of Pseudomonas aeruginosa near a vertical wall during initial attachment stages of biofilm formation
Scientific Reports (2021)
-
Mechanomicrobiology: how bacteria sense and respond to forces
Nature Reviews Microbiology (2020)
-
Re-entrant bimodality in spheroidal chiral swimmers in shear flow
Scientific Reports (2018)