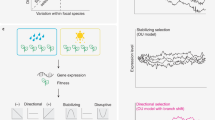
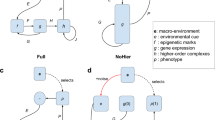
Key Points
-
Cis-regulatory elements (CREs) have essential roles in development, and their divergence is a common cause of evolutionary change.
-
Recent technological advances facilitate finding and studying orthologous CREs.
-
Functional divergence of CREs has been shown to result from small numbers of single-nucleotide substitutions, insertions and/or deletions that disrupt transcription factor binding and often interact epistatically.
-
Novel enhancer activities appear frequently to result from the co-option of transcription factor binding sites that are already present in ancestral enhancers.
-
Elucidating the specific genetic changes that are responsible for cis-regulatory divergence has provided insight into the relative contributions of new mutations and standing genetic variation, large and small effect mutations and neutral and non-neutral changes.
-
Studies of cis-regulatory divergence that underlie convergent phenotypes address questions about the repeatability of evolution.
-
Future work should focus on elucidating the relationships among sequence changes in CREs, transcription factor binding, gene expression and organismal phenotypes.
Abstract
Cis-regulatory sequences, such as enhancers and promoters, control development and physiology by regulating gene expression. Mutations that affect the function of these sequences contribute to phenotypic diversity within and between species. With many case studies implicating divergent cis-regulatory activity in phenotypic evolution, researchers have recently begun to elucidate the genetic and molecular mechanisms that are responsible for cis-regulatory divergence. Approaches include detailed functional analysis of individual cis-regulatory elements and comparing mechanisms of gene regulation among species using the latest genomic tools. Despite the limited number of mechanistic studies published to date, this work shows how cis-regulatory activity can diverge and how studies of cis-regulatory divergence can address long-standing questions about the genetic mechanisms of phenotypic evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Zuckerkandl, E. & Pauling, L. in Evolving Genes and Proteins (eds Bryson, V. & Vogel, H.) 97–166 (Academic Press, New York, 1965).
Britten, R. J. & Davidson, E. H. Gene regulation for higher cells: a theory. Science 165, 349–357 (1969).
King, M.-C. & Wilson, A. C. Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975).
Carroll, S. B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 (2008).
Stern, D. L. & Orgogozo, V. The loci of evolution: how predictable is genetic evolution? Evolution 62, 2155–2177 (2008).
Ong, C.-T. & Corces, V. G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature Rev. Genet. 12, 283–293 (2011).
Levine, M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 (2010).
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011).
Brown, R. P. & Feder, M. E. Reverse transcriptional profiling: non-correspondence of transcript level variation and proximal promoter polymorphism. BMC Genomics 6, 110 (2005).
Savinkova, L. K. et al. TATA box polymorphisms in human gene promoters and associated hereditary pathologies. Biochemistry 74, 117–129 (2009).
Wray, G. A. The evolutionary significance of cis-regulatory mutations. Nature Rev. Genet. 8, 206–216 (2007).
Hong, J. W., Hendrix, D. A. & Levine, M. S. Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 (2008).
Perry, M. W., Boettiger, A. N., Bothma, J. P. & Levine, M. Shadow enhancers foster robustness of Drosophila gastrulation. Current Biol. 20, 1562–1567 (2010).
Kleinjan, D. A. & van Heyningen, V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32 (2005).
Hare, E. E., Peterson, B. K., Iyer, V. N., Meier, R. & Eisen, M. B. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 4, e1000106 (2008).
Cande, J., Goltsev, Y. & Levine, M. S. Conservation of enhancer location in divergent insects. Proc. Natl Acad. Sci. USA 106, 14414–14419, (2009).
Kalay, G. & Wittkopp, P. J. Nomadic enhancers: tissue-specific cis-regulatory elements of yellow have divergent genomic positions among Drosophila species. PLoS Genet. 6, e1001222 (2010).
Cowles, C. R., Hirschhorn, J. N., Altshuler, D. & Lander, E. S. Detection of regulatory variation in mouse genes. Nature Genet. 32, 432–437 (2002).
Wittkopp, P. J., Haerum, B. K. & Clark, A. G. Evolutionary changes in cis and trans gene regulation. Nature 430, 85–88 (2004).
Tirosh, I., Reikhav, S., Levy, A. A. & Barkai, N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324, 659–662 (2009).
McManus, C. J. et al. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20, 816–825 (2010).
He, X., Ling, X. & Sinha, S. Alignment and prediction of cis-regulatory modules based on a probabilistic model of evolution. PLoS Comput. Biol. 5, e1000299 (2009).
Matys, V. et al. Transfac: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378 (2003).
Abdulrehman, D. et al. Yeastract: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 39, D136–D140 (2011).
Roy, S. et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 (2010).
Visel, A. et al. ChIP–seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009).
Fujita, P. A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39, D876–D882 (2011).
Hudson, M. E. Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol. Ecol. Res. 8, 3–17 (2008).
Warming, S., Costantino, N., Court, D. L., Jenkins, N. A. & Copeland, N. G. Simple and highly efficient BAC recombineering using GalK selection. Nucleic Acids Res. 33, e36 (2005).
Ghim, C. M., Lee, S. K., Takayama, S. & Mitchell, R. J. The art of reporter proteins in science: past, present and future applications. BMB Reports 43, 451–460 (2010).
Groth, A. C., Fish., M., Nusse, R. & Calos, M. P. Construction of transgenic Drosophila by using the site-specific integrase from phage PhiC31. Genetics 166, 1775–1782 (2004).
Holtzman, S. et al. Transgenic tools for members of the genus Drosophila with sequenced genomes. Fly 4, 349–362 (2010).
Gompel, N., Prud'homme, B., Wittkopp, P. J., Kassner, V. A. & Carroll, S. B. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487 (2005).
Williams, T. M. et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623 (2008).
Shirangi, T. R., Dufour, H. D., Williams, T. M. & Carroll, S. B. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 7, e1000168 (2009). This study shows how deletions in cis -regulatory sequences can lead to the formation of binding sites for transcriptional activators, resulting in divergent expression.
Deplancke, B. et al. A gene-centered C. elegans protein–DNA interaction network. Cell 125, 1193–1205 (2006).
Drewett, V. et al. DNA-bound transcription factor complexes analysed by mass-spectrometry: binding of novel proteins to the human c-fos SRE and related sequences. Nucleic Acids Res. 29, 479–487 (2001).
Stormo, G. D. & Zhao, Y. Determining the specificity of protein–DNA interactions. Nature Rev. Genet. 11, 751–760 (2010).
Berger, M. F. et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133, 1266–1276 (2008).
Maerkl, S. J. & Quake, S. R. A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007).
Frankel, N. et al. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474, 598–603 (2011). This study is one of the most detailed dissections of divergent enhancer activity to date. Divergent sites were tested individually and in combination for their effects on gene expression as well as on the divergent phenotype (loss of trichomes in D. sechellia larvae).
Jeong, S. et al. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132, 783–793 (2008).
Prabhakar, S. et al. Human-specific gain of function in a developmental enhancer. Science 321, 1346–1350, (2008).
Rebeiz, M., Jikomes, N., Kassner, V. A. & Carroll, S. B. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc. Natl Acad. Sci.USA 108, 10036–10043 (2011). This study screened tissue-specific gene expression across multiple developmental stages among closely related species of Drosophila to identify novel expression patterns. Genetic changes responsible for these putatively novel enhancer activities were then identified.
Chan, Y. F. et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (2010). This study implicates recurrent deletions of fragile cis -regulatory DNA in the repeated loss of pelvic structures in freshwater threespine stickleback.
McLean, C. Y. et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471, 216–219 (2011). This study identifies over 500 sequences that are highly conserved among chimpanzees and other mammals but that are deleted in humans. In two cases, the deletions are confirmed to remove cis -regulatory sequences and to cause changes in expression that correlate with human-specific traits.
Kidwell, M. G. & Lisch, D. Transposable elements as sources of variation in animals and plants. Proc. Natl Acad. Sci. USA 94, 7704–7711 (1997).
Daborn, P. J. et al. A single p450 allele associated with insecticide resistance in Drosophila. Science 297, 2253–2256 (2002).
Chung, H. et al. Cis-regulatory elements in the accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175, 1071–1077 (2007).
Schlenke, T. A. & Begun, D. J. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl Acad. USA 101, 1626–1631 (2004).
Bejerano, G. et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441, 87–90 (2006).
Wang, T. et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumour suppressor protein p53. Proc. Natl Acad. Sci. USA 104, 18613–18618 (2007).
Jeong, S., Rokas, A. & Carroll, S. B. Regulation of body pigmentation by the abdominal-b hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387–1399 (2006).
He, B. Z., Holloway, A. K., Maerkl, S. J. & Kreitman, M. Does positive selection drive transcription factor binding site turnover? A test with Drosophila cis-regulatory modules. PLoS Genet. 7, e1002053 (2011). This study identifies derived changes in either Drosophila melanogaster or D. simulans in 645 experimentally identified binding sites for 30 different TFs across 118 CREs and infers the effect of the derived change on TF binding affinity.
Dowell, R. D. Transcription factor binding variation in the evolution of gene regulation. Trends Genet. 26, 468–475 (2010).
Odom, D. T. et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nature Genet. 39, 730–732 (2007).
Borneman, A. R. et al. Divergence of transcription factor binding sites across related yeast species. Science 317, 815–819 (2007).
Tuch, B. B., Galgoczy, D. J., Hernday, A. D., Li, H. & Johnson, A. D. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6, e38 (2008).
Schmidt, D. et al. Five-vertebrate ChIP–seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040 (2010). This study uses ChIP coupled with next-generation sequencing to quantify the binding of two TFs in liver cells from five vertebrate species.
Kasowski, M. et al. Variation in transcription factor binding among humans. Science 328, 232–235 (2010).
Wilson, M. D. et al. Species-specific transcription in mice carrying human chromosome 21. Science 322, 434–438 (2008).
Zheng, W., Zhao, H., Mancera, E., Steinmetz, L. M. & Snyder, M. Genetic analysis of variation in transcription factor binding in yeast. Nature 464, 1187–1191 (2010).
Bradley, R. K. et al. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 8, e1000343 (2010).
Conte, C., Dastugue, B. & Vaury, C. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell. Biol. 22, 1767–1777 (2002).
Lerman, D. N., Michalak, P., Helin, A. B., Bettencourt, B. R. & Feder, M. E. Modification of heat-shock gene expression in Drosophila melanogaster populations via transposable elements. Mol. Biol. Evol. 20, 135–144 (2003).
Eichenlaub, M. P. & Ettwiller, L. De novo genesis of enhancers in vertebrates. PLoS Biol. 9, e1001188 (2011).
Prud'homme, B. et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440, 1050–1053 (2006). This study identifies the enhancers as well as localizes the specific mutations within those enhancers responsible for the independent gain of wing-specific pigmentation spots in two Drosophila species.
Sumiyama, K. & Saitou, N. Loss-of-function mutation in a repressor module of human-specifically activated enhancer HACNS1. Mol. Biol. Evol. 28, 3005–3007 (2011). This study shows that expression appearing to result from the evolution of a novel CRE in humans was more likely caused by human-specific mutations disrupting ancestral repressive sequences masking latent cis -regulatory activities.
Rebeiz, M., Pool, J. E., Kassner, V. A., Aquadro, C. F. & Carroll, S. B. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326, 1663–1667 (2009).
Babbitt, C. C. et al. Multiple functional variants in cis modulate PDYN expression. Mol Biol. Evol. 27, 465–479 (2010).
Przeworski, M. The signature of positive selection at randomly chosen loci. Genetics 160, 1179–1189 (2002).
Pool, J. E. & Aquadro, C. F. The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Mol. Ecol. 16, 2844–2851 (2007).
Wittkopp, P. J. et al. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science 326, 540–544 (2009).
Colosimo, P. F. et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 (2005).
Orr, H. A. The genetic theory of adaptation: a brief history. Nature Rev. Genet. 6, 119–127 (2005).
Mackay, T. F. Mutations and quantitative genetic variation: lessons from Drosophila. Phil. Trans. R. Soc. B 365, 1229–1239 (2010).
Charlesworth, B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nature Rev. Genet. 10, 195–205 (2009).
Arendt, J. & Reznick, D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23, 26–32 (2008).
Sucena, E., Delon, I., Jones, I., Payre, F. & Stern, D. L. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424, 935–938 (2003).
Duret, L. & Galtier, N. Comment on “human-specific gain of function in a developmental enhancer”. Science 323, 714; author reply 714 (2009).
Houle, D., Govindaraju, D. R. & Omholt, S. Phenomics: the next challenge. Nature Rev. Genet. 11, 855–866 (2010).
Hoekstra, H. E. & Coyne, J. A. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995–1016 (2007).
Lynch, V. J. & Wagner, G. P. Resurrecting the role of transcription factor change in developmental evolution. Evolution 62, 2131–2154 (2008).
Wittkopp, P. J., Haerum, B. K. & Clark, A. G. Regulatory changes underlying expression differences within and between Drosophila species. Nature Genet. 40, 346–350 (2008).
Emerson, J. J. et al. Natural selection on cis and trans regulation in yeasts. Genome Res. 20, 826–836 (2010).
Cretekos, C. J. et al. Regulatory divergence modifies limb length between mammals. Genes Dev. 22, 141–151 (2008).
Reed, R. D. et al. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333, 1137–1141 (2011).
Gomez-Skarmeta, J. L., Lenhard, B. & Becker, T. S. New technologies, new findings, and new concepts in the study of vertebrate cis-regulatory sequences. Dev. Dyn. 235, 870–885 (2006).
Richards, S. et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15, 1–18 (2005).
Peterson, B. K. et al. Big genomes facilitate the comparative identification of regulatory elements. PLoS ONE 4, e4688 (2009).
Wittkopp, P. J. Evolution of cis-regulatory sequence and function in Diptera. Heredity 97, 139–147 (2006).
Weirauch, M. T. & Hughes, T. R. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 26, 66–74 (2010).
Meireles-Filho, A. C. & Stark, A. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr. Opin. Genet. Dev. 19, 565–570 (2009).
Ludwig, M. Z., Bergman, C., Patel, N. H. & Kreitman, M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564–567 (2000).
Ludwig, M. Z. et al. Functional evolution of a cis-regulatory module. PLoS Biol. 3, e93 (2005).
Piano, F., Parisi, M. J., Karess, R. & Kambysellis, M. P. Evidence for redundancy but not trans factor—cis element co-evolution in the regulation of Drosophila Yp genes. Genetics 152, 605–616, (1999).
Wittkopp, P. J., Vaccaro, K. & Carroll, S. B. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12, 1547–1556 (2002).
Swanson, C. I., Schwimmer, D. B. & Barolo, S. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr. Biol. 21, 1186–1196 (2011).
Arnone, M. I., Dmochowski, I. J. & Gache, C. Using reporter genes to study cis-regulatory elements. Methods Cell Biol. 74, 621–652 (2004).
O'Doherty, A. et al. An aneuploid mouse strain carrying human chromosome 21 with down syndrome phenotypes. Science 309, 2033–2037 (2005).
Berman, B. P. et al. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5, R61 (2004).
Acknowledgements
This research was supported by National Science Foundation grant MCB-1021398 to P.J.W., who is an Alfred P. Sloan Research Fellow.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Selected studies identifying the genetic changes responsible for cis-regulatory divergence between species (XLS 46 kb)
Related links
Glossary
- Cis-regulatory elements
-
(CREs). Collections of transcription factor binding sites and other non-coding DNA that are sufficient to activate transcription in a defined spatial and/or temporal expression domain.
- Functionally homologous enhancers
-
Enhancers from different species that drive analogous expression patterns in the same tissue. They may or may not be caused by orthologous DNA sequences.
- Divergent sites
-
Fixed nucleotide differences in orthologous DNA sequences between species.
- Pleiotropic
-
An adjective that is used to describe a mutation or gene that affects multiple (presumably distinct) phenotypes.
- Convergent phenotypes
-
Similar phenotypes that independently evolved in different lineages.
- Trans-regulatory factors
-
Proteins, RNAs or other diffusible molecules that affect gene expression.
- Candidate sites
-
In the context of this article, one or more nucleotide changes that correlate with a change in expression and thus might be causing the change in expression.
- Degeneracy
-
In the context of transcription factor binding sites, this is the ability of a transcription factor to bind to multiple (usually related) DNA sequences.
- Outgroup
-
A related but taxonomically more distant species that can be used to infer the ancestral state of a particular site in DNA.
- Trans-regulatory environment
-
The collection of proteins, RNAs and other trans-acting molecules within a cell.
- Host species
-
In the context of transgenic analysis, the species transformed with foreign DNA.
- Position weight matrices
-
Quantitative representations of DNA-binding specificity for a transcription factor protein.
- Selective sweep
-
The increase in frequency of an allele (and closely linked chromosomal segments) that is caused by selection for the allele.
Rights and permissions
About this article
Cite this article
Wittkopp, P., Kalay, G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13, 59–69 (2012). https://doi.org/10.1038/nrg3095
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3095
This article is cited by
-
Identification and expression analysis of ATP-binding cassette (ABC) transporters revealed its role in regulating stress response in pear (Pyrus bretchneideri)
BMC Genomics (2024)
-
Maize ZmLAZ1-3 gene negatively regulates drought tolerance in transgenic Arabidopsis
BMC Plant Biology (2024)
-
Genome-wide analysis of the ABC gene family in almond and functional predictions during flower development, freezing stress, and salt stress
BMC Plant Biology (2024)
-
Genomic dissection of the correlation between milk yield and various health traits using functional and evolutionary information about imputed sequence variants of 34,497 German Holstein cows
BMC Genomics (2024)
-
Comprehensive analysis of annexin gene family and its expression in response to branching architecture and salt stress in crape myrtle
BMC Plant Biology (2024)