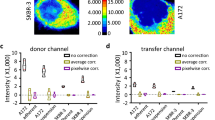
Abstract
Interactions of proteins are examined by detecting their overlap using fluorescent markers. The observed overlap is then quantified to serve as a measure of spatial correlation. A major drawback of this approach is that it can produce false values because of the properties of the image background. To remedy this, we provide a protocol to reduce the contribution of image background and then apply a protein proximity index (PPI) and correlation coefficient to estimate colocalization. Background heterogeneity is reduced by the median filtering procedure, comprising two steps, to reduce random noise and background, respectively. Alternatively, background can be reduced by advanced thresholding. PPI provides separate values for each channel to characterize the contribution of each protein, whereas correlation coefficient determines the overall colocalization. The protocol is demonstrated using computer-simulated and real biological images. It minimizes human bias and can be universally applied to various cell types in which there is a need to understand protein-protein interactions. Background reductions require 3–5 min per image. Quantifications take <1 min. The entire procedure takes approximately 15–30 min.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout














Similar content being viewed by others
References
Kuriyan, J. & Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 (2007).
Konig, P. et al. FRET-CLSM and double-labeling indirect immunofluorescence to detect close association of proteins in tissue sections. Lab. Invest. 86, 853–864 (2006).
Smallcombe, A. Multicolor imaging: the important question of co-localization. BioTechniques 30, 1240–1242, 1244–1246 (2001).
Zinchuk, V. & Grossenbacher-Zinchuk, O. Recent advances in quantitative colocalization analysis: focus on neuroscience. Prog. Histochem. Cytochem. 44, 125–172 (2009).
Manders, E.M.M., Verbeek, F.J. & Aten, J.A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993).
Bolte, S. & Cordelieres, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Zinchuk, V. & Zinchuk, O. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr. Protoc. Cell Biol. Chapter 4 Unit 4 19 (2008).
French, A.P., Mills, S., Swarup, R., Bennett, M.J. & Pridmore, T.P. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3, 619–628 (2008).
Waters, J.C. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185, 1135–1148 (2009).
Ono, M. et al. Quantitative comparison of anti-fading media for confocal laser scanning microscopy. J. Histochem. Cytochem. 49, 305–312 (2001).
Landmann, L. & Marbet, P. Colocalization analysis yields superior results after image restoration. Microsc. Res. Tech. 64, 103–112 (2004).
Zinchuk, V., Zinchuk, O., Akimaru, K., Moriya, F. & Okada, T. Ethanol consumption alters expression and colocalization of bile salt export pump and multidrug resistance protein 2 in the rat. Histochem. Cell Biol. 127, 503–512 (2007).
Shaw, P. & Rawlins, D.J. The point spread function of confocal microscope: its measurement and use in deconvolution. J. Microsc. 163, 151–165 (1991).
Wu, Y. et al. Quantitative determination of spatial protein-protein correlations in fluorescence confocal microscopy. Biophys. J. 98, 493–504 (2010).
Lippincott-Schwartz, J. & Manley, S. Putting super-resolution fluorescence microscopy to work. Nat. Methods 6, 21–23 (2009).
Haugland, R.P. The Handbook: A Guide to Fluorescent Probes and Labeling Technologies 10th edn (Invitrogen, Inc., 2005).
Schneider Gasser, E.M. et al. Immunofluorescence in brain sections: simultaneous detection of presynaptic and postsynaptic proteins in identified neurons. Nat. Protoc. 1, 1887–1897 (2006).
Adler, J. & Parmryd, I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A 77, 733–742 (2010).
Acknowledgements
This work was partially supported by National Institutes of Health grant no. HL088640 and the American Heart Association Postdoctoral Fellowship no. 10POST4230081. We thank M. Celio (Fribourg University) for help in organizing the study.
Author information
Authors and Affiliations
Contributions
V.Z. conceived and organized the study, designed and conducted experiments and wrote the paper; Y.W. conducted experiments and contributed to writing, O.G.-Z. designed and conducted experiments and contributed to writing; and E.S. helped to organize the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Pair 1: 0% colocalization. Red channel. (TIFF 769 kb)
Supplementary Fig. 2
Pair 1: 0% colocalization. Green channel. (TIFF 769 kb)
Supplementary Fig. 3
Pair 2: 25% colocalization. Red channel. (TIFF 769 kb)
Supplementary Fig. 4
Pair 2: 25% colocalization. Green channel. (TIFF 769 kb)
Supplementary Fig. 5
Pair 3: 50% colocalization. Red channel. (TIFF 769 kb)
Supplementary Fig. 6
Pair 3: 50% colocalization. Green channel. (TIFF 769 kb)
Supplementary Fig. 7
Pair 4: 75% colocalization. Red channel. (TIFF 769 kb)
Supplementary Fig. 8
Pair 4: 75% colocalization. Green channel. (TIFF 769 kb)
Rights and permissions
About this article
Cite this article
Zinchuk, V., Wu, Y., Grossenbacher-Zinchuk, O. et al. Quantifying spatial correlations of fluorescent markers using enhanced background reduction with protein proximity index and correlation coefficient estimations. Nat Protoc 6, 1554–1567 (2011). https://doi.org/10.1038/nprot.2011.384
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.384
This article is cited by
-
Techniques for detecting protein-protein interactions in living cells: principles, limitations, and recent progress
Science China Life Sciences (2019)
-
Variable-angle epifluorescence microscopy characterizes protein dynamics in the vicinity of plasma membrane in plant cells
BMC Plant Biology (2018)
-
MatCol: a tool to measure fluorescence signal colocalisation in biological systems
Scientific Reports (2017)
-
TSC but not PTEN loss in starving cones of retinitis pigmentosa mice leads to an autophagy defect and mTORC1 dissociation from the lysosome
Cell Death & Disease (2016)
-
Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria
Nature (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.