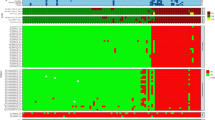
Abstract
The mouse monoclonal antibody (mAb) technology still represents a key source of reagents for research and clinical diagnosis, although it is relatively inefficient and expensive and therefore unsuitable for high-throughput production against a vast repertoire of antigens. In this article, we describe a protocol that combines the immunization of individual mice with complex mixtures of influenza virus strains and a microarray-based immunoassay procedure to perform a parallel screening against the viral antigens. The protocol involves testing the supernatants of somatic cell hybrids against a capture substratum containing an array of different antigens. For each fusion experiment, we carried out more than 25,000 antigen-antibody reactivity tests in less than a week, a throughput that is two orders of magnitude higher than that of traditional antibody detection assays such as enzyme-linked immunosorbent assays and immunofluorescence. Using a limited number of mice, we can develop a vast repertoire of mAbs directed against nuclear and surface proteins of several human and avian influenza virus strains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kohler, G. & Milstein, C. Continuous cultures of fused cells secreting antibody of predetermined specificity. Nature 256, 495–497 (1975).
Wengelnik, K. et al. The A-domain and the thrombospondin-related motif of Plasmodium falciparum TRAP are implicated in the invasion process of mosquito salivary glands. EMBO J. 18, 5195–5204 (1999).
Davis, J.M., Pennington, J.E., Kubler, A.-M. & Conscience, J.F. A simple, single-step technique for selecting and cloning hybridomas for the production of monoclonal antibodies. J. Immunol. Methods 50, 161–171 (1982).
Ayriss, J., Valero, R., Bradbury, A.R. & Pavlik, P. Multiplexed flow cytometry: high-throughput screening of single-chain antibodies. Methods Mol. Biol. 525, 241–60 (2009).
Seideman, J. & Peritt, D. A novel monoclonal antibody screening method using the Luminex-100k microsphere system. J. Immunol. Methods 267, 165–171 (2002).
Quesniaux, V., Himmelspach, K. & van Regenmortel, M.H. An enzyme immunoassay for the screening of monoclonal antibodies to cyclosporine. Immunol. Lett. 9, 99–104 (1985).
Hubbard, R. & Wiseman, A. Enzyme immunoassay and the use of monoclonal antibodies. Trends Analyt. Chem. 2, VII–IX (1983).
Vignali, D.A.A. Multiplexed particle-based flow cytometric assays. J. Immunol. Methods 243, 243–255 (2000).
Ayriss, J., Woods, T., Bradbury, A. & Pavlik, P. High-throughput screening of single-chain antibodies using multiplexed flow cytometry. J. Proteome Res. 6, 1072–1082 (2007).
Sasaki, K., Glass, T.R. & Ohmura, N. Validation of accuracy of enzyme-linked immunosorbent assay in hybridoma screening and proposal of an improved screening method. Anal. Chem. 77, 1933–1939 (2005).
Taylor, P. Optimizing assays for automated platforms. Mod. Drug Discov. 5, 37–39 (2002).
Liu, B., Li, S. & Hu, J. Technological advances in high-throughput screening. Am. J. Pharmacogenomics 4, 263–76 (2004).
Fernandes, P.B. Technological advances in high-throughput screening. Curr. Opin. Chem. Biol. 2, 597–603 (1998).
Taipa, M.A. Immunoassays: biological tools for high throughput screening and characterisation of combinatorial libraries. Comb. Chem. High Throughput Screen. 11, 325–335 (2008).
Joelsson, D., Moravec, P., Troutman, M., Pigeon, J. & DePhillips, P. Optimizing ELISAs for precision and robustness using laboratory automation and statistical design of experiments. J. Immunol. Methods 337, 35–41 (2008).
Bacarese-Hamilton, T., Gray, J. & Crisanti, A. Protein microarray technology for unraveling the antibody specificity repertoire against microbial proteomes. Curr. Opin. Mol. Ther. 5, 278–284 (2003).
Ardizzoni, A. et al. A protein microarray immunoassay for the serological evaluation of the antibody response in vertically transmitted infections. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1067–1075 (2009).
Bacarese-Hamilton, T., Ardizzoni, A., Gray, J. & Crisanti, A. Protein arrays for serodiagnosis of disease. Methods Mol. Biol. 264, 271–283 (2004).
Ogunniyi, A.O., Story, C.M., Papa, E., Guillen, E. & Love, J.C. Screening individual hybridomas by microengraving to discover monoclonal antibodies. Nat. Protoc. 4, 767–782 (2009).
De Masi, F. et al. High throughput production of mouse monoclonal antibodies using antigen microarrays. Proteomics 5, 4070–4081 (2005).
Bacarese-Hamilton, T., Mezzasoma, L., Ardizzoni, A., Bistoni, F. & Crisanti, A. Serodiagnosis of infectious diseases with antigen microarrays. J. Appl. Microbiol. 96, 10–17 (2004).
Mezzasoma, L., Bacarese-Hamilton, T., Di Cristina, M., Rossi, R., Bistoni, F. & Crisanti, A. Antigen microarrays for serodiagnosis of infectious diseases. Clin. Chem. 48, 121–130 (2002).
Bacarese-Hamilton, T., Bistoni, F. & Crisanti, A. Protein microarrays: from serodiagnosis to whole proteome scale analysis of the immune response against pathogenic microorganisms. Biotechniques Dec (Suppl), 24–29 (2002).
Bacarese-Hamilton, T. et al. Detection of allergen-specific IgE on microarrays by use of signal amplification techniques. Clin. Chem. 48, 1367–1370 (2002).
Bacarese-Hamilton, T., Gray, J., Ardizzoni, A. & Crisanti, A. Allergen microarrays. Methods Mol. Med. 114, 195–207 (2005).
Gray, J.C. et al. Profiling the antibody immune response against blood stage malaria vaccine candidates. Clin. Chem. 53, 1244–1253 (2007).
MacBeath, G. & Schreiber, S.L. Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 (2000).
Larsson, K., Wester, K., Nilsson, P., Uhlén, M., Hober., S. & Wernérus, H. Multiplexed PrEST immunization for high-throughput affinity proteomics. J. Immunol. Methods 315, 110–120 (2006).
Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. Appendix 3B, 1–2 (2001).
Acknowledgements
We thank I. Donatelli for providing the influenza viruses and antigens used in all studies. This work was supported by a grant from EU FP & Health FLUARRAY (GA n 201960) and by FIRB grants RBIP064CRT and RBLA03C9F4.
Author information
Authors and Affiliations
Contributions
M.D. designed the experiments, analyzed data, supervised the project and wrote the paper; L.N., M.A.G. and B.C. conducted the experiments and analyzed the data; L.D., G.M., F.B. and R.S. intellectually contributed to this work; A.C. inspired and supervised the project, and wrote and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Comparison of array reactivity using slides from different batches. The images show arrays processed using the same supernatant (MAb 2A12 in panel a and MAb 7G11 in panel b, see Supplementary Table 1) but printed at different times onto different batches of slides. (TIFF 3319 kb)
Supplementary Figure 2
Images of whole slides containing several identical arrays. Each slide contains four replicas of the same array processed with the supernatants of the hybridomas 1H11, 11D7, 22C12, 17G11 (see Supplementary Table 1). The same reactivity patter observed in all array replicas on one slide demonstrates the very low variability among intra-slide spots. (TIFF 4457 kb)
Supplementary Figure 3
Layout of samples in the 384 well plate and array design. a) Table reporting the order of samples in the wells of the 384 microplate. The spotting buffer used to print each sample is reported in the right column. b) Window of the MicroGrid II software displaying the layout of the 384 microplate. A schematic representation of the 384 microplate is shown on the bottom of the panel b. The twenty-eight wells loaded with the spotting samples have been numbered. c) This screen enables the desired pin array pattern to be entered. The array pattern is designed by entering the number of the 384 microplate well in the editor. d) This screen enables one to define the position of the tool-array. By adjusting horizontal (top and bottom) and vertical (left and right) margins the pin tool placement area can be altered and the arrays that fit are generated automatically. (TIFF 4073 kb)
Supplementary Figure 4
MAb reacting to glycosyl residues. Microarray (left panel) and immunoblot (right panel) reactivity profile of the MAb 1H11 reacting against distantly related influenza virus types. The immunoblot reactivity of the MAb 1H11 was analysed against both glycosylated and de-glycosylated (Deg) virus preparations. Removal of glycosyl residues was performed using the Enzymatic Protein De-glycosylation Kit (Sigma). (TIFF 3314 kb)
Supplementary Table 1
Microarray reactivity profile against array influenza antigens of the selected MAbs. (XLS 40 kb)
Rights and permissions
About this article
Cite this article
Di Cristina, M., Nunziangeli, L., Giubilei, M. et al. An antigen microarray immunoassay for multiplex screening of mouse monoclonal antibodies. Nat Protoc 5, 1932–1944 (2010). https://doi.org/10.1038/nprot.2010.161
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.161
This article is cited by
-
Surface-imprinted polymers in microfluidic devices
Science China Chemistry (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.