Abstract
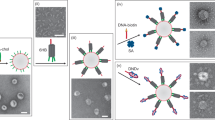
Artificial lipid-bilayer membranes are valuable tools for the study of membrane structure and dynamics. For applications such as the study of vesicular transport and drug delivery, there is a pressing need for artificial vesicles with controlled size. However, controlling vesicle size and shape with nanometre precision is challenging, and approaches to achieve this can be heavily affected by lipid composition. Here, we present a bio-inspired templating method to generate highly monodispersed sub-100-nm unilamellar vesicles, where liposome self-assembly was nucleated and confined inside rigid DNA nanotemplates. Using this method, we produce homogeneous liposomes with four distinct predefined sizes. We also show that the method can be used with a variety of lipid compositions and probe the mechanism of templated liposome formation by capturing key intermediates during membrane self-assembly. The DNA nanotemplating strategy represents a conceptually novel way to guide lipid bilayer formation and could be generalized to engineer complex membrane/protein structures with nanoscale precision.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cho, W. & Stahelin, R. V. Membrane–protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 34, 119–151 (2005).
Rigaud, J.-L. & Lévy, D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372, 65–86 (2003).
Bally, M. et al. Liposome and lipid bilayer arrays towards biosensing applications. Small 6, 2481–2497 (2010).
Allen, T. M. & Cullis, P. R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 65, 36–48 (2013).
Çağdaş, M., Sezer, A. D. & Bucak, S. Liposomes as Potential Drug Carrier Systems for Drug Delivery (InTech, 2014).
Dua, J., Rana, A. & Bhandari, A. Liposome: methods of preparation and applications. Int. J. Pharm. Stud. Res. 3, 14–20 (2012).
Gregory, G. Liposome Technology Vol. 1 (CRC, 2006).
Batzri, S. & Korn, E. D. Single bilayer liposomes prepared without sonication. Biochim. Biophys. Acta 298, 1015–1019 (1973).
Bosworth, M. E., Hunt, C. A. & Pratt, D. Liposome dialysis for improved size distributions. J. Pharm. Sci. 71, 806–812 (1982).
Mayer, L. D., Hope, M. J., Cullis, P. R. & Janoff, A. S. Solute distributions and trapping efficiencies observed in freeze-thawed multilamellar vesicles. Biochim. Biophys. Acta 817, 193–196 (1985).
Pidgeon, C., McNeely, S., Schmidt, T. & Johnson, J. E. Multilayered vesicles prepared by reverse-phase evaporation: liposome structure and optimum solute entrapment. Biochemistry 26, 17–29 (1987).
Genc, R., Ortiz, M. & O'Sullivan, C. K. Curvature-tuned preparation of nanoliposomes. Langmuir 25, 12604–12613 (2009).
Mouritsen, O. G. Lipids, curvature, and nano-medicine. Eur. J. Lipid Sci. Technol. 113, 1174–1187 (2011).
Sun, J. et al. Tunable rigidity of (polymeric core)–(lipid shell) nanoparticles for regulated cellular uptake. Adv. Mater. 27, 1402–1407 (2015).
Schubert, R. Liposome preparation by detergent removal. Methods Enzymol. 367, 46–70 (2003).
Holzer, M., Barnert, S., Momm, J. & Schubert, R. Preparative size exclusion chromatography combined with detergent removal as a versatile tool to prepare unilamellar and spherical liposomes of highly uniform size distribution. J. Chromatogr. A 1216, 5838–5848 (2009).
Huang, C. Studies on phosphatidylcholine vesicles. Formation and physical characteristics. Biochemistry 8, 344–352 (1969).
Olson, F., Hunt, C. A., Szoka, F. C., Vail, W. J. & Papahadjopoulos, D. Preparation of liposomes of defined size distribution by extrusion through polycarbonate membranes. Biochim. Biophys. Acta 557, 9–23 (1979).
Hope, M. J., Bally, M. B., Webb, G. & Cullis, P. R. Production of large unilamellar vesicles by a rapid extrusion procedure: characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta 812, 55–65 (1985).
Silva, R., Ferreira, H., Little, C. & Cavaco-Paulo, A. Effect of ultrasound parameters for unilamellar liposome preparation. Ultrason. Sonochem. 17, 628–632 (2010).
Wang, T., Wang, N., Wang, T., Sun, W. & Li, T. Preparation of submicron liposomes exhibiting efficient entrapment of drugs by freeze-drying water-in-oil emulsions. Chem. Phys. Lipids 164, 151–157 (2011).
Yu, D. G. et al. Self-assembled liposomes from amphiphilic electrospun nanofibers. Soft Matter 7, 8239–8247 (2011).
Jahn, A., Lucas, F., Wepf, R. A. & Dittrich, P. S. Freezing continuous-flow self-assembly in a microfluidic device: toward imaging of liposome formation. Langmuir 29, 1717–1723 (2013).
McMahon, H. T. & Gallop, J. L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596 (2005).
Langecker, M., Arnaut, V., List, J. & Simmel, F. C. DNA nanostructures interacting with lipid bilayer membranes. Acc. Chem. Res. 47, 1807–1815 (2014).
Beales, P. A. & Vanderlick, T. K. Specific binding of different vesicle populations by the hybridization of membrane-anchored DNA. J. Phys. Chem. A 111, 12372–12380 (2007).
Chan, Y. H., van Lengerich, B. & Boxer, S. G. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc. Natl Acad. Sci. USA 106, 979–984 (2009).
Dave, N. & Liu, J. Programmable assembly of DNA-functionalized liposomes by DNA. ACS Nano 5, 1304–1312 (2011).
Pinheiro, A. V., Han, D., Shih, W. M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nature Nanotech. 6, 763–772 (2011).
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Nanomaterials. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Li, P. et al. A pH-sensitive multifunctional gene carrier assembled via layer-by-layer technique for efficient gene delivery. Int. J. Nanomed. 7, 925–939 (2012).
Dong, Y. et al. Frame-guided assembly of vesicles with programmed geometry and dimensions. Angew. Chem. Int. Ed. 53, 2607–2610 (2014).
Perrault, S. D. & Shih, W. M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 8, 5132–5140 (2014).
Lin, C., Perrault, S. D., Kwak, M., Graf, F. & Shih, W. M. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 41, e40 (2013).
Kourembanas, S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 77, 13–27 (2015).
Vinson, P. K., Talmon, Y. & Walter, A. Vesicle–micelle transition of phosphatidylcholine and octyl glucoside elucidated by cryo-transmission electron-microscopy. Biophys. J. 56, 669–681 (1989).
Cho, H.-J. et al. Measurement of ice thickness on vitreous ice embedded cryo-EM grids: investigation of optimizing condition for visualizing macromolecules. J. Anal. Sci. Technol. 4, 7 (2013).
Ludtke, S. J. & Chiu, W. Focal pair merging for contrast enhancement of single particles. J. Struct. Biol. 144, 73–78 (2003).
Kaler, E. W., Murthy, A. K., Rodriguez, B. E. & Zasadzinski, J. A. Spontaneous vesicle formation in aqueous mixtures of single-tailed surfactants. Science 245, 1371–1374 (1989).
Li, W., Huang, Z., MacKay, J. A., Grube, S. & Szoka, F. C. Jr. Low-pH-sensitive poly(ethylene glycol) (PEG)-stabilized plasmid nanolipoparticles: effects of PEG chain length, lipid composition and assembly conditions on gene delivery. J. Gene Med. 7, 67–79 (2005).
Li, W. & Szoka, F. C. Jr. Lipid-based nanoparticles for nucleic acid delivery. Pharm. Res. 24, 438–449 (2007).
Ollivon, M., Lesieur, S., Grabielle-Madelmont, C. & Paternostre, M. Vesicle reconstitution from lipid–detergent mixed micelles. Biochim. Biophys. Acta 1508, 34–50 (2000).
Stuart, M. C. & Boekema, E. J. Two distinct mechanisms of vesicle-to-micelle and micelle-to-vesicle transition are mediated by the packing parameter of phospholipid–detergent systems. Biochim. Biophys. Acta 1768, 2681–2689 (2007).
Mimms, L. T., Zampighi, G., Nozaki, Y., Tanford, C. & Reynolds, J. A. Phospholipid vesicle formation and transmembrane protein incorporation using octyl glucoside. Biochemistry 20, 833–840 (1981).
Zumbuehl, O. & Weder, H. G. Liposomes of controllable size in the range of 40 to 180 nm by defined dialysis of lipid/detergent mixed micelles. Biochim. Biophys. Acta 640, 252–262 (1981).
Philippot, J., Mutaftschiev, S. & Liautard, J. P. A very mild method allowing the encapsulation of very high amounts of macromolecules into very large (1000 nm) unilamellar liposomes. Biochim. Biophys. Acta 734, 137–143 (1983).
Iinuma, R. et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344, 65–69 (2014).
Gerling, T., Wagenbauer, K. F., Neuner, A. M. & Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015).
Acknowledgements
The authors thank P.D. Ellis for designing the DNA rings used for placing lipid seeds at different angles and for proofreading the manuscript, and F. Sigworth for providing cryo-electron micrographs of extruded liposomes. This work is supported by a National Institutes of Health (NIH) Director's New Innovator Award (DP2-GM114830), an NIH grant (R21-GM109466) and a Yale University faculty startup fund to C.L., an NIH grant to J.E.R. (R01-DK027044) and an NIH Director's New Innovator Award (DP2-OD004641), an Army Research Office MURI grant (W911NF-12-1-0420), National Science Foundation Expeditions Grant (1317694) and a Wyss Institute for Biologically Inspired Engineering Faculty Award to W.M.S.
Author information
Authors and Affiliations
Contributions
Y.Y. and J.W. designed and conducted the majority of the experiments, analysed the data, prepared the majority of the manuscript and contributed equally to this work. H.S. performed cryo-EM studies and prepared the manuscript. W.X. initiated the project, developed the DNA–lipid conjugation method and designed and performed pilot experiments to prepare and purify DNA-ring enclosed vesicles. W.M.S. initiated the project and discussed the results. J.E.R. initiated the project, supervised J.W. and W.X., and discussed the results. C.L. initiated the project, designed and supervised the study, interpreted the data and prepared the manuscript. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 47625 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Wang, J., Shigematsu, H. et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nature Chem 8, 476–483 (2016). https://doi.org/10.1038/nchem.2472
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2472
This article is cited by
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)
-
Digital nanoreactors to control absolute stoichiometry and spatiotemporal behavior of DNA receptors within lipid bilayers
Nature Communications (2023)
-
Preparation, applications, and challenges of functional DNA nanomaterials
Nano Research (2023)
-
Functionalizing DNA origami to investigate and interact with biological systems
Nature Reviews Materials (2022)
-
Wrap to sort
Nature Chemistry (2021)