Abstract
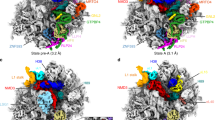
Eukaryotic ribosomes consist of a small 40S and a large 60S subunit that are assembled in a highly coordinated manner. More than 200 factors ensure correct modification, processing and folding of ribosomal RNA and the timely incorporation of ribosomal proteins1,2. Small subunit maturation ends in the cytosol, when the final rRNA precursor, 18S-E, is cleaved at site 3 by the endonuclease NOB13. Previous structures of human 40S precursors have shown that NOB1 is kept in an inactive state by its partner PNO14. The final maturation events, including the activation of NOB1 for the decisive rRNA-cleavage step and the mechanisms driving the dissociation of the last biogenesis factors have, however, remained unresolved. Here we report five cryo-electron microscopy structures of human 40S subunit precursors, which describe the compositional and conformational progression during the final steps of 40S assembly. Our structures explain the central role of RIOK1 in the displacement and dissociation of PNO1, which in turn allows conformational changes and activation of the endonuclease NOB1. In addition, we observe two factors, eukaryotic translation initiation factor 1A domain-containing protein (EIF1AD) and leucine-rich repeat-containing protein 47 (LRRC47), which bind to late pre-40S particles near RIOK1 and the central rRNA helix 44. Finally, functional data shows that EIF1AD is required for efficient assembly factor recycling and 18S-E processing. Our results thus enable a detailed understanding of the last steps in 40S formation in human cells and, in addition, provide evidence for principal differences in small ribosomal subunit formation between humans and the model organism Saccharomyces cerevisiae.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Woolford, J. L. Jr & Baserga, S. J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013).
Bohnsack, K. E. & Bohnsack, M. T. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 38, e100278 (2019).
Henras, A. K., Plisson-Chastang, C., O’Donohue, M.-F., Chakraborty, A. & Gleizes, P.-E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225–242 (2015).
Ameismeier, M., Cheng, J., Berninghausen, O. & Beckmann, R. Visualizing late states of human 40S ribosomal subunit maturation. Nature 558, 249–253 (2018).
Cerezo, E. et al. Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip. Rev. RNA 10, e1516 (2019).
Strunk, B. S., Novak, M. N., Young, C. L. & Karbstein, K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150, 111–121 (2012).
Lebaron, S. et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 19, 744–753 (2012).
Turowski, T. W. et al. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 42, 12189–12199 (2014).
Widmann, B. et al. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell 23, 22–35 (2012).
Angermayr, M., Roidl, A. & Bandlow, W. Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol. Microbiol. 44, 309–324 (2002).
Ferreira-Cerca, S., Kiburu, I., Thomson, E., LaRonde, N. & Hurt, E. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 42, 8635–8647 (2014).
Laronde-LeBlanc, N., Guszczynski, T., Copeland, T. & Wlodawer, A. Structure and activity of the atypical serine kinase Rio1. FEBS J. 272, 3698–3713 (2005).
Meyer, B. et al. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res. 44, 4304–4316 (2016).
Babaian, A. et al. Loss of m1acp3Ψ ribosomal RNA modification is a major feature of cancer. Cell Rep. 31, 107611 (2020).
Hector, R. D. et al. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 42, 12138–12154 (2014).
Heuer, A. et al. Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae. eLife 6, e30189 (2017).
Scaiola, A. et al. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J. 37, e98499 (2018).
O’Donohue, M. F., Choesmel, V., Faubladier, M., Fichant, G. & Gleizes, P. E. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 190, 853–866 (2010).
Montellese, C. et al. USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit. eLife 9, e54435 (2020).
Badertscher, L. et al. Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep. 13, 2879–2891 (2015).
Farley-Barnes, K. I. et al. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep. 22, 1923–1934 (2018).
Yu, J. & Marintchev, A. Comparative sequence and structure analysis of eIF1A and eIF1AD. BMC Struct. Biol. 18, 11 (2018).
Bertomeu, T. et al. A high-resolution genome-wide CRISPR/Cas9 viability screen reveals structural features and contextual diversity of the human cell-essential proteome. Mol. Cell. Biol. 38, e00302-17 (2018).
Acker, M. G., Shin, B. S., Dever, T. E. & Lorsch, J. R. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 281, 8469–8475 (2006).
Zemp, I. et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185, 1167–1180 (2009).
Pertschy, B. et al. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 284, 35079–35091 (2009).
Sloan, K. E., Knox, A. A., Wells, G. R., Schneider, C. & Watkins, N. J. Interactions and activities of factors involved in the late stages of human 18S rRNA maturation. RNA Biol. 16, 196–210 (2019).
Lamanna, A. C. & Karbstein, K. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc. Natl Acad. Sci. USA 106, 14259–14264 (2009).
Espinar-Marchena, F. J., Babiano, R. & Cruz, J. Placeholder factors in ribosome biogenesis: please, pave my way. Microb. Cell 4, 144–168 (2017).
Wyler, E. et al. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA 17, 189–200 (2011).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Taoka, M. et al. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 46, 9289–9298 (2018).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Zemp, I. et al. CK1δ and CK1ε are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J. Cell Sci. 127, 1242–1253 (2014).
van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733 (2019).
Acknowledgements
We thank S. Rieder, C. Ungewickell, H. Sieber and A. Gilmozzi for technical assistance, T. Fröhlich for mass-spectrometry analysis and L. Kater, J. Cheng and T. Becker for discussions and critical comments on the manuscript. This project has received funding from the Deutsche Forschungsgemeinschaft (DFG) through the GRK1721 to R.B., a DFG fellowship through the QBM (Quantitative Biosciences Munich) graduate school to M.A., from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 885711—Human-Ribogenesis) to R.B., and the Swiss National Science Foundation (the NCCR ‘RNA and disease’ and grant 31003A_166565) to U.K.
Author information
Authors and Affiliations
Contributions
M.A., R.B., I.Z., J.v.d.H. and U.K designed the study. M.A. and M.T. generated stable cell lines and purified native complexes. M.A. and M.T. prepared the cryo-EM samples and O.B. collected cryo-EM data. M.A. processed the cryo-EM data and built the molecular models. For the functional analysis of novel RBFs, I.Z. and J.v.d.H. performed cellular analyses. I.Z., J.v.d.H. and U.K. analysed the data and interpreted results. All authors interpreted the combined results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Sebastian Klinge, David Tollervey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
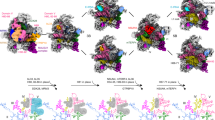
Extended Data Fig. 1 Sample preparation and cryo-EM data analysis.
a, SDS–PAGE analysis of native pre-40S complexes purified with RIOK1(D324) and NOB1(D10N). Identified protein bands are labelled. For gel source data, see Supplementary Fig. 1. b, Representative micrographs from the three data sets. Scale bar 50 nm c, Subset of 2D averages of extracted particles after initial 2D classification. d, e, Summarized classification scheme of RIOK1(D324A) (d) and NOB1(D10N) (e). Particles of final states, marked in orange, were subjected to CTF parameter refinement and Bayesian polishing before the last 3D refinement. f, Particles of state G from both NOB1(D10N) data sets were combined and classified. Final volume is marked in orange. g, Summarized classification scheme of LRRC47.
Extended Data Fig. 2 Local resolution, refinement and model statistics.
a, Local resolution distribution of states F1 – H2 with their respective colour grading scheme as estimated by Relion. b, Fourier shell correlation (FSC) curve for all states. Average resolution values as stated in Fig. 1 are calculated according to the ‘gold standard’ at FSC = 0.143. c, FSC plot of the models against their volume as provided by Phenix. d, Local resolution estimation of the ‘head’ region of state F2 after focused refinement. e, f, Model and cryo-EM density of state H1 around the post-transcriptionally modified rRNA residues C1337, G1490 (e), and A1832 (see ref. 43.) (f).
Extended Data Fig. 3 Details on RIOK1 binding to the pre-40S particle.
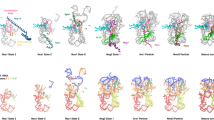
a, Superposition of models of RIOK1 and RIOK2 after alignment of state D (PDB-6G51) and H1 highlight the overlapping binding site at the decoding centre and the rotation of their central RIO domain by approximately 15°. b, Cartoon representation of eL41 (PDB-6G5H), as well as the C terminus of LTV1 (PDB-6G51) and RIOK1 relative to the top of the matured h44 in state H1. Overlaps in binding sites highlight mutually exclusive binding. c, Models of state F1, F2 and H1 with focus on RIOK1 position. Unaccounted density within the mRNA entry tunnel in states F1 and F2 (yellow) is likely the flexible N terminus of RIOK1, which overlaps with the two helices of factor X present in state E (marked with a red cross, see ref. 4.). d, Surface representation of state D (PDB-6G51) with RBF TSR1 in cartoon representation. Models of RIOK1 (left) and LRRC47 (right) of state F1 after alignment of the particles emphasize the overlaps in binding sites around h44. e, Post-transcriptional modification of U1248 leads to formation of 1-methyl-3-α-amino-α-carboxypropyl pseudouridine (m1acp3Ψ). Modifying enzymes and their contribution to the structure are indicated by colours. f, Cartoon representation showing the coordination of m1acp3Ψ1248 by RIOK1.
Extended Data Fig. 4 Structural details of novel factors LRRC47 and EIF1AD.
a, Overall structure of the two domains of LRRC47. Leucine residues, secondary structure elements and position of h44 are highlighted. b, Low-resolution cryo-EM reconstruction of a sample using N-terminally tagged LRRC47 (left) and state H1 filtered at 7 Å (right). The leucine-rich domain of LRRC47 (blue) binds simultaneously with TSR1, while the C-terminal domain remains delocalized and is therefore not visible. c, Models of LRRC47, h44 and parts of TSR1 show the conformational changes in h44 that accompany the transition between the states. A central part of h44 moves after release of TSR1, enabled by the lack of its N-terminal helix (left). The B3/4 domain of LRRC47 would clash with both displayed helices of TSR1. LRRC47 continues to bind in an almost unchanged position after maturation of h44 (right). d, Structure of EIF1AD with its N-terminal helix and residues N36, R58, K59 and W62 labelled. e, Cartoon representation of rRNA segments and ribosomal proteins surrounding EIF1AD in state H1. Model of yeast eIF1A (PDB-6GSN) after alignment of a pre-48S translation initiation complex to the pre-40S particle shows a shifted binding location. f, Sequence alignment of human EIF1AD and eIF1A. Conserved residues that bind to rRNA are coloured blue and the IDDI motif of eIF1A in red. The conserved C-terminal stretch that binds to uS13 and uS19 is marked with a blue box. g, Model of state H1 with cryo-EM volume of EIF1AD and eS25 Gaussian filtered at 1.5 standard deviations. Additional density extends from well resolved parts of the C terminus of EIF1AD. Detailed views on eS25 N terminus and EIF1AD C terminus are shown in boxes A and B.
Extended Data Fig. 5 EIF1AD but not LRRC47 depletion affects late 40S subunit maturation.
a, Western blot analysis of the experiment shown in Fig. 4 confirming the effectiveness of siRNA treatments for EIF1AD and RIOK2. b, Western blot analysis for the experiment in c confirming the depletion of LRRC47 or RIOK2 upon siRNA treatment. For gel source data of a and b, see Supplementary Fig. 1. c, Immunofluorescence analysis of HeLa cells treated with siRNAs against LRRC47 or RIOK2 using antibodies against the indicated RBFs. For immunofluorescence analysis of NOB1, cells were treated with 20 nM leptomycin B (LMB) for 90 min. Note that only RIOK2 but not LRRC47 depletion leads to cytoplasmic recycling defects of the tested RBFs. Scale bar, 20 μm. d, FISH analysis of experiment in b, revealing cytoplasmic accumulation of 18S-E pre-rRNA upon RIOK2 but not LRRC47 depletion. FISH pictures were processed in parallel, using a gamma correction of 1.5. All experiments were done in triplicates (n = 3).
Extended Data Fig. 6 Structural details of rRNA 3′end maturation.
a, Cartoon representation of the 3′ end of 18S-E rRNA with PNO1, NOB1 and eS26 throughout the maturation process in states F2, G and H1. Panel of eS26 with 18S rRNA in state H1 has been shifted slightly upwards as indicated by the line to the right. b, Detailed view of NOB1(D10N) active site with its substrate. Dashed circle marks site 3 cleavage site. Electron density around ITS1 and the 3′end shown in blue. c, Stick representation of NOB1 residues that interact with the ITS1 and 3′ end with electron density shown in blue.
Supplementary information
Supplementary Figure
Supplementary Figure 1 This file contains a figure with the uncropped SDS-PAGE and western blot images. Related to Extended Data Figs. 1 and 5.
Rights and permissions
About this article
Cite this article
Ameismeier, M., Zemp, I., van den Heuvel, J. et al. Structural basis for the final steps of human 40S ribosome maturation. Nature 587, 683–687 (2020). https://doi.org/10.1038/s41586-020-2929-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2929-x
This article is cited by
-
Domain definition and preliminary functional exploration of the endonuclease NOBP-1 in Strongyloides stercoralis
Parasites & Vectors (2023)
-
Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles
Cell Research (2023)
-
Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2
Nature Communications (2023)
-
PNO1 regulates autophagy and apoptosis of hepatocellular carcinoma via the MAPK signaling pathway
Cell Death & Disease (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.