Abstract
N6-methyladenosine (m6A) is the most prevalent modified nucleotide in mRNA1,2, with around 25% of mRNAs containing at least one m6A. Methylation of mRNA to form m6A is required for diverse cellular and physiological processes3. Although the presence of m6A in an mRNA can affect its fate in different ways, it is unclear how m6A directs this process and why the effects of m6A can vary in different cellular contexts. Here we show that the cytosolic m6A-binding proteins—YTHDF1, YTHDF2 and YTHDF3—undergo liquid–liquid phase separation in vitro and in cells. This phase separation is markedly enhanced by mRNAs that contain multiple, but not single, m6A residues. Polymethylated mRNAs act as a multivalent scaffold for the binding of YTHDF proteins, juxtaposing their low-complexity domains and thereby leading to phase separation. The resulting mRNA–YTHDF complexes then partition into different endogenous phase-separated compartments, such as P-bodies, stress granules or neuronal RNA granules. m6A-mRNA is subject to compartment-specific regulation, including a reduction in the stability and translation of mRNA. These studies reveal that the number and distribution of m6A sites in cellular mRNAs can regulate and influence the composition of the phase-separated transcriptome, and suggest that the cellular properties of m6A-modified mRNAs are governed by liquid–liquid phase separation principles.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA sequencing (Fig. 4e, Extended Data Fig. 5a) and ribosome profiling (Fig. 4f, g, Extended Data Fig. 5b, d) data reported in this paper have been deposited in the NCBI Gene Expression Omnibus under accession number GSE125725. All other data are available from the corresponding author upon reasonable request.
References
Perry, R. P. & Kelley, D. E. Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42 (1974).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971–3975 (1974).
Meyer, K. D. & Jaffrey, S. R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127 (2018).
Nott, T. J. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Quiroz, F. G. & Chilkoti, A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 14, 1164–1171 (2015).
Wiśniewski, J. R., Hein, M. Y., Cox, J. & Mann, M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics 13, 3497–3506 (2014).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382(2017).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Kroschwald, S. et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807 (2015).
Standart, N. & Weil, D. P-bodies: cytosolic droplets for coordinated mRNA storage. Trends Genet. 34, 612–626 (2018).
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Anders, M. et al. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1, e201800113 (2018).
Slobodin, B., Han, R., Calderone, V., Vrielink, J. A., Loayza-Puch, F., Elkon, R. & Agami, R. Transcription impacts the efficiency of mRNA translation via co-transcriptional N 6-adenosine methylation. Cell 169, 326–337.e312 (2017).
Knuckles, P. et al. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat. Struct. Mol. Biol. 24, 561–569 (2017).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Wang, X. et al. Structural basis of N 6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016).
Śledź, P. & Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 5, e18434 (2016).
Han, T. W. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 (2012).
Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R. & Parker, R. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820.e805 (2017).
Namkoong, S., Ho, A., Woo, Y. M., Kwak, H. & Lee, J. H. Systematic characterization of stress-induced RNA granulation. Mol. Cell 70, 175–187.e178 (2018).
Wang, X. et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Bouche, G., Amalric, F., Caizergues-Ferrer, M. & Zalta, J. P. Effects of heat shock on gene expression and subcellular protein distribution in Chinese hamster ovary cells. Nucleic Acids Res. 7, 1739–1747 (1979).
Van Treeck, B. & Parker, R. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell 174, 791–802 (2018).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Kedersha, N. et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 (2005).
Zhu, T. et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N 6-methyladenosine. Cell Res. 24, 1493–1496 (2014).
Xiang, Y. et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017).
Geula, S. et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Knuckles, P. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 (2018).
Wen, J. et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e1026 (2018).
Patil, D. P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Xu, C. et al. Structural basis for the discriminative recognition of N 6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290, 24902–24913 (2015).
Zhang, H. et al. RNA controls polyQ protein phase transitions. Mol. Cell 60, 220–230 (2015).
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008).
Hoover, D. M. & Lubkowski, J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 30, e43 (2002).
Li, H. et al. Design and specificity of long ssDNA donors for CRISPR-based knock-in. Preprint at https://www.biorxiv.org/content/10.1101/178905v1 (2017).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
McGlincy, N. J. & Ingolia, N. T. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129 (2017).
Xiao, Z., Zou, Q., Liu, Y. & Yang, X. Genome-wide assessment of differential translations with ribosome profiling data. Nat. Commun. 7, 11194 (2016).
Mi, H. et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210 (2010).
Lancaster, A. K., Nutter-Upham, A., Lindquist, S. & King, O. D. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502 (2014).
Gilles, J. F., Dos Santos, M., Boudier, T., Bolte, S. & Heck, N. DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 115, 55–64 (2017).
Hubstenberger, A. et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157.e145 (2017).
Jain, S. et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016).
Youn, J. Y. et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e511 (2018).
Markmiller, S. et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e513 (2018).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576.e516 (2017).
Acknowledgements
We thank members of the Jaffrey laboratory for comments and suggestions; members of the Epigenomics, Optical Microscopy and Imaging & Flow Cytometry Weill Cornell Cores for their assistance; and J. Hanna and S. Geula for generously providing Mettl14-knockout and control mES cell lines. This work was supported by NIH grants R01DA037755 (S.R.J.), F32CA22104-01 (B.F.P.), R01DK114131 (J.H.L.) and T32CA062948 (A.-O.G.) and an American-Italian Cancer Foundation fellowship (S.Z.).
Peer review information
Nature thanks Richard Kriwacki and Tanja Mittag for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
S.R.J., R.J.R., A.O.-G., S.Z. and P.K. designed the experiments. R.J.R. and P.K. carried out stress-granule-staining experiments; S.N., J.H.L. and H.K. prepared stress granules; R.J.R. and P.K. performed assays related to phase separation and stress-granule formation; P.K. performed puromycin-labelling assays, R.J.R. performed quantification of stress granules and smFISH; R.J.R., P.K. and S.Z. analysed ribosome-profiling data; A.O.-G. and R.J.R. performed analysis of stress-granule transcriptomes; B.F.P. performed m6A measurements and D.P.P. made DF expression constructs. S.Z. performed CRISPR knock-in; R.J.R. and S.Z. performed in-cell FRAP experiments. R.J.R. and S.Z. prepared figures relating to ribosome-profiling data. R.J.R., S.Z. and P.K. prepared the remaining figures. S.R.J. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
S.R.J. is scientific founder of, advisor to, and owns equity in Gotham Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Fluorescent labelling of DF2 does not affect the formation of liquid droplets.
a, DF1, DF2 and DF3 exhibit high sequence homology. Shown is a colour-coded schematic representation of the aligned amino acid sequence and corresponding prion-like domain disorder propensity plots (red and black traces) for DF1, DF2 and DF3 generated using the PLAAC (prion-like amino acid composition) tool46. The y axis of the plot represents prion-like regions (1) and regions of background amino acid composition (0). The low-complexity domain is a region of approximately 40 kDa that contains glutamine-rich prion-like domains and an abundance of disorder-promoting residues such as proline, glycine, serine, alanine and asparagine. These domains are also enriched with multiple P-Xn-G motifs that are known to be associated with lower critical solution temperature7. The YTH domain (about 15 kDa) exhibits high sequence identity between the paralogues, and all YTH domains show identical binding to m6A without preference for any specific sequence context surrounding m6A4. The high degree of sequence identity suggests that these proteins might function redundantly in stress-granule formation and phase separation. Amino acid compositions of the full-length DF proteins and their prion-like domains are shown in the bar charts at the bottom of the panel. b, Liquid-droplet formation for Alexa488-labelled DF2. The goal of this experiment is to confirm that labelling DF2 with Alexa488 does not affect liquid-droplet formation. Indeed, before labelling DF2 with Alexa488, DF2 protein droplets were readily detectable by differential interference contrast microscopy (left). After labelling, Alexa488-labelled DF2 protein droplets are still observed by fluorescence microscopy (right). These data indicate that the labelling protocol does not impair droplet formation by DF2. Images are taken from different protein preparations. Experiments were performed in duplicate. Scale bar, 10 µm. c, The intrinsically disordered domain of DF2 is required for the phase separation of DF2. Bright-field microscopic images of recombinant DF2 lacking the N-terminal intrinsically ordered domain (YTH) and full length DF2 are shown (a schematic of the domain representation is shown above the image). The edge of the buffer (buffer/air interface) is shown with a dashed line. Whereas the full-length YHTDF2 (75 µM) can undergo phase separation, at the same concentration and in the same buffer conditions YTH cannot. This indicates that the intrinsically disordered domain is required for phase separation. Experiments were performed in duplicate. Scale bar, 10 µm. d, DF1 and DF3 undergo phase separation in vitro. Shown are fluorescence microscopy images of Alexa594–DF1 and Alexa647–DF3. DF1 and DF3 undergo phase separation in vitro as assessed by the formation of protein droplets. Experiments were performed in duplicate. Scale bar, 10 µm. e, DF1, DF2 and DF3 form protein droplets comprising all three proteins. Shown are fluorescence microscopy images of Alexa594–DF1, Alexa488–DF2 and Alexa647–DF3. Mixing the three recombinant proteins shows that these proteins can phase-separate together to form protein droplets that contain all three proteins. Experiments were performed in duplicate. Scale bar, 10 µm. f, Confirmation of in vitro-transcribed RNA abundance and methylation status. In vitro-transcribed RNAs were serially diluted (1:10) and stained for total RNA by methylene blue staining (top left) as well as m6A abundance by immunoblotting using an anti-m6A antibody (bottom left). RNA with no m6A nucleotides gave no signal whereas RNAs with 10 m6A nucleotides gave a significantly higher signal in the dot blot than those with 1 m6A nucleotide. Additionally, in vitro-transcribed RNAs were analysed on a 15% denaturing gel, demonstrating the absence of degradation products (right). Experiments were performed in duplicate. g, Partition coefficients of fluorescently labelled m6A RNAs with and without DF2. To determine the extent to which multi-m6A-RNAs were recruited into DF droplets, we synthesized a 10-m6A RNA with a 5′ BODIPY FL fluorescent tag and measured its partition coefficient in the presence of DF2 (7.5 µM, 20 mM HEPES pH 7.4, 300 mM KCl, 6 mM MgCl2, 0.02% NP-40, 10% glycerol). Upon addition of 850 nM BODIPY–10-m6A-RNA, fluorescent RNA-containing droplets appeared in minutes (left). A video of fluorescent DF2:BODIPY–10-m6A-RNA coacervate droplet fusion is shown in Supplementary Video 2. Calculation of partition coefficients in comparison to background fluorescent-labelled RNAs6 shows that m6A mRNAs are enriched in DF2-containing droplets (right; RNA only, n = 11; RNA + DF2, n = 24, where n represents distinct droplets in biological replicates). The experiment was performed in duplicate. Bar heights represent mean partition coefficients and error bars represent s.e.m. ****P < 0.0001, two-sided Mann–Whitney test. Scale bar, 10 µm. h, The partition coefficient of DF proteins increases over time. In this experiment we measured the partition coefficient of DF1, DF2 and DF3 as shown in Fig. 1g. However, here we measured the values after 24 h, unlike the time point used in Fig. 1g (approximately 5 min). The partition coefficients are notably increased compared to the values measured in Fig. 1g. This suggests that droplet formation had not achieved equilibrium at the early time points used in Fig. 1g. Bar heights represent mean partition coefficients and error bars represent s.e.m. Experiments were performed in duplicate.
Extended Data Fig. 2 Assessing which stressors induce stress granules and the localization of DF2 proteins in diverse cell types.
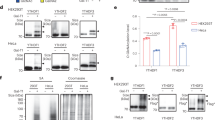
a, Oxidative stress and heat shock induce stress-granule formation in mouse ES cells. Stress-granule formation has not been extensively characterized in mouse ES cells. We therefore wanted to ensure that stress granule composition is the same in mouse ES cells compared to other cell types in which stress granules are more frequently studied. To test mouse ES stress granules, we stained with additional markers. Co-immunostaining with ATXN2 (green) and G3BP1 (red) after arsenite treatment (0.5 mM for 1 h) and heat shock (42 °C for 30 min) in mES cells showed clear labelling of stress granules. The overlay panel shows ATXN2 and G3BP1 overlap (yellow). Thus, stress granules in mouse ES cells appear to have similar markers as stress granules in other cell types. The experiment was performed in triplicate. Scale bar, 10 μm. b, c, DF1 and DF3 proteins relocalize to stress granules after heat shock and oxidative stress. DF1, DF2 and DF3 have high sequence similarity and show similar phase-separation properties. We therefore wanted to determine whether all these proteins associate with stress granules. Co-immunostaining was performed in mES cells with DF1 (red) or DF3 (red) with TIAR (green) after arsenite treatment (0.5 mM for 1 h) or heat shock (42 °C for 30 min). Along with DF2 shown in Fig. 2, DF1 and DF3 relocalize to stress granules treatment as visualized by the co-localization with TIAR. Scale bar, 10 µm. These findings are consistent with previous proteomic datasets of stress granules. A P-body proteome dataset48 showed that DF2 was enriched in P-bodies. DF2 ranked 152 among 1,900 P-body-associated proteins by abundance. All DF proteins were identified in a group of around 300 stress-granule-enriched proteins in a proteomics study of stress granules49. In another study, in vivo proximity-dependent biotinylation (BioID)-labelling study of G3BP1 and other stress-granule markers showed interactions with all DF proteins50. Another APEX labelling study51 of G3BP1 showed that the YTHDFs are 3 of the top 42 G3BP1-interacting proteins in the stress-granule proteome. Overall, these studies suggest that DF proteins are commonly seen in stress granules, and may be highly abundant relative to other stress-granule components. The experiment was performed in triplicate. d–f, DF2 relocalizes to stress granules after arsenite treatment in numerous cell types. The focus of this experiment was to determine whether DF relocalization to stress granules is likely to be a universal feature of stress granules. We therefore tested DF localization to stress granules in multiple cell types. Shown is co-immunostaining of HEK293 cells (d), U2OS cells (e), and NIH3T3 cells (f) with DF2 (red) and TIAR (green) after arsenite treatment (0.5 mM for 1 h) and heat shock (42 °C for 30 min). The overlay panel shows DF2 in stress granules based on its overlap with TIAR (yellow). The experiment was performed in duplicate. Scale bar, 10 μm. g, Confirmation of CRISPR–Cas9 knock in of NeonGreen–DF2. A western blot of HEK293T shows endogenous expression of NeonGreen–DF2. Note, only one allele contains the knock-in construct, accounting for the presence of unmodified DF2 in cells. h, Arsenite stress induces the localization of NeonGreen–DF2 into stress granules. We wanted to determine whether the ability of DF2 to undergo phase separation in vitro could be actively observed in cells. Unstressed HEK293T cells expressing NeonGreen-tagged DF2 protein show a diffuse cytoplasmic fluorescent signal. Upon arsenite stress (0.5 mM, 1 h), NeonGreen–DF2 phase-separates into stress granules. This confirms the ability of NeonGreen–DF2 to undergo phase separation in cells in response to stress. The experiment was performed in triplicate. Scale bar, 10 μm. i, Relocalization of DF2 to the nucleus does not occur after various stresses in various cell types. Because DF2 has been reported to relocalize to the nucleus 2 h after heat shock13, we wanted to determine whether any nuclear relocalization occurs in our experiments, which were performed immediately after stress. The ‘Stress condition’ column indicates the type and length of stress applied. The ‘Cell type’ column indicates the type of cell that was stressed. The ‘DF2 in nucleus’ column denotes the number of cells that were found to have DF2 in the nucleus immediately after stress. The ‘Total cells’ column indicates the number of cells that were examined for DF2 nuclear relocalization in each experimental condition. In all conditions, there was no cell that showed nuclear DF2 localization. Thus, DF2 localization is primarily in cytosolic stress granules at the time at which the stress is terminated. DF2 was not observed to relocalize to the nucleus at any time point or after any stress, including the 2-h post-heat-shock conditions described previously13. j, DF2 relocalization to stress granules does not require new mRNA or protein synthesis. We wanted to know whether an increase in DF2 expression or new m6A formation could be required for the formation of stress granules after heat shock. To test this, we blocked protein synthesis with puromycin and blocked new transcription with actinomycin D. Actinomycin D blocks m6A formation because m6A formation occurs co-transcriptionally17,18. The fluorescence micrographs show DF2 immunostaining in HEK293T cells treated with DMSO (left), puromycin (10 μg ml−1, middle), and actinomycin D (2.5 μg ml−1, right) for 15 min before and during incubation at 42 °C for 30 min. The ability of DF2 to relocalize to stress granules when transcription (actinomycin D) and translation (puromycin) was arrested was assessed by immunofluorescence staining for DF2. In each case, the formation of stress granules was unaffected, indicating that no new protein synthesis or new methylation is required for stress-granule formation. The time course of stress-granule formation is rapid, making it unlikely that new protein synthesis or methylation is involved in stress-granule formation. Additionally, heat shock is normally associated with inhibited transcription and translation, which further suggests that new protein synthesis and RNA methylation is unlikely to occur in the time course of stress-granule formation. On the basis of all this data, we propose that stress granule formation probably utilizes pre-existing patterns of m6A seen in unstressed cells to mediate the formation of stress granules. The experiment was performed in duplicate. Scale bar, 10 µm. k, l, m6A levels are not significantly altered immediately after arsenite and heat-shock stress in NIH3T3 cells. We wanted to test whether m6A levels in mRNA transcripts were altered as a result of cellular stress. NIH3T3 cells were subjected to arsenite (0.5 mM, 1 h) or heat-shock stress (43 °C, 45 min) and total RNA was extracted immediately after stress treatment. Total RNA was further purified by poly(A) selection to specifically assay m6A levels in mRNA transcripts. TLC38 revealed that there was no significant increase in m6A levels within poly(A) mRNA immediately after either stress condition in three biological replicates (see l). This indicates that cellular stress does not induce an increase or decrease in m6A over the time frame examined. Experiments were performed in duplicate. Bar heights in l represent mean and error bars represent s.e.m. Three biological replicates (n = 3) were analysed in the control, and four biological replicates (n = 4) were analysed after heat-shock and arsenite stress. Stress m6A/(A+C+U) ratios were analysed with a two-sided Student’s t-test.
Extended Data Fig. 3 Confirmation of the Mettl14-knockout model and DF2 phase-separation into P-bodies in mES cells.
a, Mettl14-knockout (KO) mES cells are depleted in m6A RNA. We sought to independently confirm the depletion of m6A from mRNA in these cells, which were previously shown to have a 99% reduction in m6A32. The TLC assay selectively quantifies m6A in a G–A–C context, thereby reducing the possibility of contamination of m6A from ribosomal RNA or small nuclear RNA, which are quantified in an A–A–C or a C–A–G context, respectively38. The protocol was performed as described previously38. Indicated in the TLC chromatograms are the relative positions of m6A (dotted circle) and adenosine (A), cytosine (C) and uracil (U). The left and right panels show radiochromatograms obtained from 2D-TLC of poly(A) RNA from wild-type and Mettl14-knockout cells. No m6A is detectable in the poly(A) RNA derived from Mettl14-knockout cells, which confirms the efficiency of m6A depletion in these cells. Experiments were performed in duplicate. mES cells are used here because m6A depletion can be readily achieved in Mettl14-knockout mES cells without impairing viability32. By contrast, m6A depletion cannot be readily achieved in immortalized cell lines as both Mettl3 and Mettl14 are essential for nearly all cell lines52. b, DF2 partitioning into stress granules induced by arsenite is delayed in m6A-deficient cells. This delay is similar to that shown in stress granules induced by heat shock as seen in Fig. 3a. The experiment was performed in triplicate.
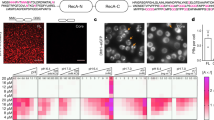
Extended Data Fig. 4 The number of m6A nucleotides is correlated with stress-granule enrichment independent of transcript length.
a, DF2 is enriched in stress granules after stress. Nuclear (Nuc), cytosolic (Cyt), insoluble RNA-granule enriched (SG, red boxed lanes), and soluble (Sol) protein fractions were isolated from stressed NIH3T3 cells as described previously24. G3BP1 was used as a stress-granule marker. GAPDH and tubulin were used as cytosolic and soluble fraction markers. Under non-stressed conditions, DF2 is most abundant in the cytoplasmic and soluble protein fractions. However, upon both arsenite and heat-shock stress, the highest levels of DF2 are found in the RNA granule (insoluble) fraction, which indicates that diverse stresses cause the partitioning of DF2 from the cytosol into stress granules. The experiment was performed in duplicate. b, m6A levels are increased in the mRNAs in the insoluble stress-granule-enriched fraction after cellular stress in NIH3T3 cells. Shown are representative TLC plates analysing m6A levels in mRNAs in the stress-granule fraction from the analysis presented in Fig. 4a. Representative plates from the cytosolic fraction are shown in Extended Data Fig. 2l. Experiments were performed in duplicate. c, m6A number correlates with mRNA enrichment in RNA granules in mouse neurons. In these experiments, we used mRNA enrichment data (RNA granule versus supernatant) derived from a study of isoxazole-induced RNA granules in mouse brain22. Enriched mRNAs are defined by a log2-transformed fold change of more than 1 in the RNA granule fraction relative to the supernatant fraction. A cumulative distribution plot of mRNA enrichment was performed for mRNAs classified by the number of called m6A peaks per gene based on single-nucleotide resolution m6A maps generated in the mouse brain. Transcripts that contain multiple m6A peaks are enriched in RNA granules relative to non-methylated or singly methylated mRNAs. The original experiments were performed in triplicate. d, The number of m6A sites in an mRNA correlates with its enrichment in stress granules in NIH3T3 cells. In these experiments, we used a dataset of relative mRNA enrichment data (stress granule versus cytoplasm) generated in a previous study24. Assignment of the number of m6A sites in each transcript was based on a mouse embryonic fibroblast MeRIP-seq dataset obtained previously32. Analysis was performed as in Fig. 4c. Polymethylated mRNAs show greater enrichment in stress granules than non-methylated or singly methylated mRNAs for each stress condition. Experiments were performed in triplicate. e, Examination of the effect of m6A on mRNA enrichment using controlled transcript size. Because transcript length positively correlates with stress-granule mRNA enrichment (see refs 23,24), we wanted to control for this feature in our analysis. The same m6A maps and RNA-seq data from U2OS cells that were used to generate Fig. 4c were used here. Transcripts were binned on the basis of their annotated transcript length (2–3 kb, 3–4 kb, 4–5 kb, 5–6 kb) and further sorted on the basis of the number of annotated m6A sites in each transcript. We found that mRNAs annotated with fixed lengths each showed increased enrichment based on the number of mapped m6A sites. The number of m6A per transcript was a positive predictor of transcript enrichment in stress granules even when controlling for transcript length. The centre of the box plot represents the median log2-transformed fold change, the boundaries contain genes within a quartile of the median, the whiskers represent genes in the upper and lower quartiles, and the outliers are presented as dots.
Extended Data Fig. 5 Detection of translation in mES cells after stress.
a, m6A-mRNA transcript abundance is similar before stress and after stress. We wanted to understand whether mRNA transcript abundance was altered as a result of DF mobilization in mES cells after heat shock. In Fig. 4a, we examined RNA expression before heat shock and compared it to mRNA levels after 30 min of heat shock. Here, we allowed the cells to recover for 1 h, reasoning that this additional time might allow for DF-mediated mRNA degradation. As in Fig. 4a, we performed RNA-seq on wild-type mES cells before heat shock and after stress, measured after cells were returned to 37 °C for 1 h. The same m6A annotation strategy was used as in Fig. 4a. As can be seen, the levels of m6A in an mRNA is not correlated with an alteration in mRNA abundance. The log2-transformed fold change values represent the average of four biological replicates. b, Raw counts for ribosome protected fragments. Ribosome-protected fragments were collected from mES cells before stress, immediately after heat shock (42 °C, 30 min) and 1 h after heat shock. The number of ribosome-protected fragments isolated from cells immediately after heat shock was substantially lower than the number of ribosome-protected fragments isolated before stress and 1 h after stress. This indicates that translation is globally suppressed during the heat shock. As a result of the few ribosome-protected fragments during heat shock, translational efficiency could not be calculated during heat shock. Bar heights represent the totals from four biological replicates in each condition. c, Translation recovers 1 h after heat shock in mES cells. Here we assessed the amount of time needed for translation to be detected after heat shock. mES cells were subject to heat shock for 30 min at 42 °C and translation was assessed at different time points after the cessation of heat shock. Translation was monitored by labelling nascent peptides with puromycin. Puromycin was added to cells for 10 min. Immunostaining with an antibody against TIAR and puromycin provides a correlation between the presence of stress granules and the translation state. Non-stressed cells that were not treated with puromycin are shown as a control to establish the background signal (upper left). Unstressed cells treated with puromycin show robust translation (green). At a recovery time of 30 min, most cells still contain stress granules (TIAR, red) and translation is absent except in the few cells that lack stress granules. However, at 1 h, translating cells can be readily detected on the basis of puromycin immunoreactivity reactivity. Less than 50% of cells exhibit stress granules. On the basis of these experiments, we used 1 h as the time point for our ribosome-profiling experiments. The experiments were performed in duplicate. Scale bar, 10 µm. d, Comparison of the two biological replicates with the highest percentage of CDS-mapped reads in each condition for ribosome-profiling experiments. Shown are Pearson’s correlation plots for the replicates used in the translational efficiency analysis shown in Fig. 4f, g.
Extended Data Fig. 6 Gene ontology of polymethylated and singly methylated mRNAs in U2OS cells.
a, b, Gene ontology of m6A-mRNAs in U2OS cells. U2OS RNA-seq data from ref. 23 and Me-RIP-seq data from ref. 31 were used in a gene ontology analysis for polymethylated m6A-mRNAs in U2OS cells. Polymethylated mRNAs were defined as all mRNAs that had four or more annotated m6A sites in the MeRIP-seq dataset that were identified in the U2OS RNA-seq (n = 652). Singly methylated mRNAs were defined as mRNAs that had one annotated m6A site, with the same criteria (n = 2,896). Gene ontology was performed using the PANTHER gene ontology database. The biological process gene ontology search showed enrichment of regulatory and developmental-associated genes in the polymethylated group. The molecular function gene ontology search showed enrichment of protein, ion, enzyme and adenylyl-binding proteins, and de-enrichment of ribosome structural components. The cellular component gene ontology search showed de-enrichment of mitochondrial and ribosomal proteins, and enrichment of components of the nucleus and cell membrane. mRNAs that met the same inclusion criteria but had no annotated m6A sites were used as the reference category for the gene ontology analysis (n = 5,956). Fold enrichment scores for each gene ontology category are indicated by the coloured bars and correspond to the left y axis. P values for each gene ontology category are indicated by the dark grey diamonds and correspond to the right y axis. P values were determined with Fisher’s exact test and a Bonferroni correction was performed for multiple hypothesis testing.
Extended Data Fig. 7 Model of how the properties of m6A-containing mRNAs are determined by their phase separation into intracellular phase-separated compartments.
a, Depicted is the binding of DF proteins to singly methylated mRNAs. DF proteins show low-affinity interactions with m6A-containing mRNAs. Affinities typically range between 0.9–1.1 µM for DF1, DF2 and DF34,36. These low affinities suggest that DF proteins would not be able to form a stable bimolecular interaction with singly methylated RNA. The low affinity can now be understood in the context of phase separation. Their weak interactions with RNA are probably stabilized by interactions between their low-complexity domains, and subsequent phase separation. Notably, all m6A sites in cytosolic mRNAs appear to have an equal propensity to bind each DF protein35. Thus, any m6A residue may be sufficient to enhance the phase-separation potential of an mRNA. However, higher-level information—such as the spacing of m6A sites—as well as other mRNA-bound proteins with low-complexity domains, may further affect the efficiency of phase separation. b, Polymethylated mRNAs bind multiple DF proteins leading to phase separation. When multiple DF proteins bind to a polymethylated mRNA, their interactions with the mRNA are stabilized by DF–DF interactions mediated by their low-complexity domains. These complexes may be reversible and undergo an assembly–disassembly equilibrium. However, if P-bodies, neuronal granules or stress granules are present, the DF–mRNA complexes can partition into these structures. m6A-mRNA is then regulated by the regulatory proteins and functional properties of these distinct structures. If an mRNA has a single m6A site, the mRNA can still partition into phase-separated structures, especially if RNA–RNA interactions or other RNA–protein interactions can facilitate phase separation. The DF low-complexity domain could interact with these non-DF proteins to enhance mRNA partitioning. Overall, interactions between DF proteins and m6A-mRNAs probably lower the saturation concentration for their incorporation within stress granules, enhancing their partitioning over non-methylated mRNAs.
Supplementary information
Supplementary Table 1
Differential expression analysis of RNA-seq data from heat shocked (42 °C, 30 min) mES cells immediately after stress and following 1 h recovery compared to unstressed cells. Analysis was performed using the DESeq2 package in R. The Wald test was used to generate p-values. A Benjamini-Hochberg correction was applied to the adjusted p-values. Data were generated from four biological replicates in each condition.
Supplementary Table 2
Translational efficiency analysis of Ribo-seq data from heat shocked (42 °C, 30 min) WT and Mettl14 KO mES cells before and after 1 h recovery from stress. Analysis was performed using the xtail package in R. Two analysis pipelines are used to generate initial p-values: “v1” p-values are generated by comparing mRNA and RPF log2 fold changes; “v2” p-values are generated by comparing log2 ratios between mRNA and RPFs. “Final” p-values for translational efficiency select the more conservative (higher) p-value from the “v1” or “v2” method. A Benjamini-Hochberg correction was applied to the adjusted p-values. For the unstressed condition, four and three biological replicates were used for WT and Mettl14 KO mES cells, respectively. For the 1 h recovery condition, four and two biological replicates were used for WT and Mettl14 KO mES cells, respectively.
Video 1
Time-lapse imaging of Alexa488-DF2 liquid droplets. Recombinant DF2 labeled with Alexa488 forms mobile liquid droplets that undergo rapid fusion. Shown is a time period during which multiple droplet fusion events can be observed (white arrowheads). Droplet fusion videos were taken in the same conditions as shown in the still images in Fig. 1d (75 µM DF2, 20 mM HEPES pH 7.4, 300 mM KCl, 6 mM MgCl2, 0.02% NP-40). Experiments were performed in triplicate. Scale bar, 10 µm.
Video 2
Time-lapse imaging of fluorescently labeled 10-m6A transcripts and DF2 RNA-protein coacervates. To visualize RNA:DF2 protein coacervates in real-time, we synthesized 10-m6A RNAs with a 5’ BODIPY FL fluorescent tag and added 850 nM of BODIPY-10-m6A-RNA to DF2 in solution (7.5 µM, 20 mM HEPES pH 7.4, 300 mM KCl, 6 mM MgCl2, 0.02% NP-40, 10% glycerol). Droplet fusion videos were taken under the same conditions as shown in the still image in Extended Data 1g. Shown is a time period during which droplet fusion events can be observed (white arrowheads). Experiments were performed in duplicate. Scale bar, 10 µm.
Rights and permissions
About this article
Cite this article
Ries, R.J., Zaccara, S., Klein, P. et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019). https://doi.org/10.1038/s41586-019-1374-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1374-1
This article is cited by
-
Decoding epitranscriptomic regulation of viral infection: mapping of RNA N6-methyladenosine by advanced sequencing technologies
Cellular & Molecular Biology Letters (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
Mex-3 RNA binding family member A (MEX3A)/circMPP6 complex promotes colorectal cancer progression by inhibiting autophagy
Signal Transduction and Targeted Therapy (2024)
-
N6-methyladenosine-mediated feedback regulation of abscisic acid perception via phase-separated ECT8 condensates in Arabidopsis
Nature Plants (2024)
-
Regulation of stress granule formation in human oligodendrocytes
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.