Abstract
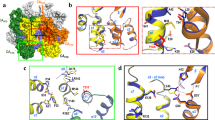
Retroviral capsid proteins are conserved structurally but assemble into different morphologies1. The mature human immunodeficiency virus-1 (HIV-1) capsid is best described by a ‘fullerene cone’ model2,3, in which hexamers of the capsid protein are linked to form a hexagonal surface lattice that is closed by incorporating 12 capsid-protein pentamers. HIV-1 capsid protein contains an amino-terminal domain (NTD) comprising seven α-helices and a β-hairpin4,5, a carboxy-terminal domain (CTD) comprising four α-helices6,7, and a flexible linker with a 310-helix connecting the two structural domains8. Structures of the capsid-protein assembly units have been determined by X-ray crystallography9,10; however, structural information regarding the assembled capsid and the contacts between the assembly units is incomplete. Here we report the cryo-electron microscopy structure of a tubular HIV-1 capsid-protein assembly at 8 Å resolution and the three-dimensional structure of a native HIV-1 core by cryo-electron tomography. The structure of the tubular assembly shows, at the three-fold interface11, a three-helix bundle with critical hydrophobic interactions. Mutagenesis studies confirm that hydrophobic residues in the centre of the three-helix bundle are crucial for capsid assembly and stability, and for viral infectivity. The cryo-electron-microscopy structures enable modelling by large-scale molecular dynamics simulation, resulting in all-atom models for the hexamer-of-hexamer and pentamer-of-hexamer elements as well as for the entire capsid. Incorporation of pentamers results in closer trimer contacts and induces acute surface curvature. The complete atomic HIV-1 capsid model provides a platform for further studies of capsid function and for targeted pharmacological intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Accessions
Protein Data Bank
Data deposits
Cryo-EM structural data have been deposited in the EMDataBank under accession codesEMD-5582 andEMD-5639, and the MDFF atomic model of the CA HOH and models of HIV-1 capsid have been deposited in the Protein Data Bank under accession numbers 3J34, 3J3Q, 3J3Y.
References
Sundquist, W. I. & Krausslich, H. G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2, a006924 (2012)
Ganser, B. K., Li, S., Klishko, V. Y., Finch, J. T. & Sundquist, W. I. Assembly and analysis of conical models for the HIV-1 core. Science 283, 80–83 (1999)
Li, S., Hill, C. P., Sundquist, W. I. & Finch, J. T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 (2000)
Gitti, R. K. et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273, 231–235 (1996)
Momany, C. et al. Crystal structure of dimeric HIV-1 capsid protein. Nature Struct. Biol. 3, 763–770 (1996)
Du, S. et al. Structure of the HIV-1 full-length capsid protein in a conformationally trapped unassembled state induced by small-molecule binding. J. Mol. Biol. 406, 371–386 (2011)
Gamble, T. R. et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278, 849–853 (1997)
Jiang, J. et al. The interdomain linker region of HIV-1 capsid protein is a critical determinant of proper core assembly and stability. Virology 421, 253–265 (2011)
Pornillos, O. et al. X-ray structures of the hexameric building block of the HIV capsid. Cell 137, 1282–1292 (2009)
Pornillos, O., Ganser-Pornillos, B. K. & Yeager, M. Atomic-level modelling of the HIV capsid. Nature 469, 424–427 (2011)
Byeon, I. J. et al. Structural convergence between cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell 139, 780–790 (2009)
Meng, X., Zhao, G. & Zhang, P. Structure of HIV-1 capsid assemblies by cryo-electron microscopy and iterative helical real-space reconstruction. J. Vis. Exp. 54, e3041 (2011)
Egelman, E. H. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 157, 83–94 (2007)
Ganser-Pornillos, B. K., Cheng, A. & Yeager, M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131, 70–79 (2007)
Trabuco, L. G., Villa, E., Schreiner, E., Harrison, C. B. & Schulten, K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49, 174–180 (2009)
Byeon, I. J. et al. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: implications for structural polymorphism of CA assemblies. J. Am. Chem. Soc. 134, 6455–6466 (2012)
Forshey, B. M., von Schwedler, U., Sundquist, W. I. & Aiken, C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76, 5667–5677 (2002)
Joshi, A., Nagashima, K. & Freed, E. O. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation. J. Virol. 80, 7939–7951 (2006)
Meng, X. et al. Protease cleavage leads to formation of mature trimer interface in HIV-1 capsid. PLoS Pathog. 8, e1002886 (2012)
Chen, B. & Tycko, R. Simulated self-assembly of the HIV-1 capsid: protein shape and native contacts are sufficient for two-dimensional lattice formation. Biophys. J. 100, 3035–3044 (2011)
Yeager, M. Design of in vitro symmetric complexes and analysis by hybrid methods reveal mechanisms of HIV capsid assembly. J. Mol. Biol. 410, 534–552 (2011)
Cardone, G., Purdy, J. G., Cheng, N., Craven, R. C. & Steven, A. C. Visualization of a missing link in retrovirus capsid assembly. Nature 457, 694–698 (2009)
Caspar, D. L. & Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962)
Briggs, J. A., Wilk, T., Welker, R., Krausslich, H. G. & Fuller, S. D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22, 1707–1715 (2003)
Bharat, T. A. et al. Structure of the immature retroviral capsid at 8 A resolution by cryo-electron microscopy. Nature 487, 385–389 (2012)
Mastronarde, D. N. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J. Microsc. 230, 212–217 (2008)
Agulleiro, J. I. & Fernandez, J. J. Fast tomographic reconstruction on multicore computers. Bioinformatics 27, 582–583 (2011)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
Acknowledgements
We thank P. Schwerdtfeger for access to his software for creating fullerene models, T. Brosenitsch for reading the manuscript, and M. DeLucia and J. Mehrens for technical assistance. This work was supported by the National Institutes of Health (GM082251, GM085043 and GM104601) and the National Science Foundation (PHY0822613, MCB0744057). Large-scale molecular dynamics simulations were performed on the Blue Waters Computer, financed by the National Science Foundation (OCI 07-25070).
Author information
Authors and Affiliations
Contributions
G.Z., J.R.P., E.L.Y., A.M.G., K.S., C.A. and P.Z. designed the research. J.N. and J.A. prepared samples for electron microscopy. G.Z. collected cryo-EM data. G.Z., X.M. and P.Z. analysed cryo-EM and cryo-ET data. E.L.Y. and C.A. performed biochemical and functional analysis. K.S. developed large-scale modelling methodology; J.R.P. performed molecular dynamics simulations and B.C. performed CG-MC simulations. G.Z., J.R.P., K.S. and P.Z. analysed atomic models. G.Z., J.R.P., A.M.G., K.S., C.A. and P.Z wrote the paper with support from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Table 1, Supplementary Figures 1-8 and Supplementary References. (PDF 7830 kb)
Molecular dynamics flexible fitting of CA assembly
Helvetica (MOV 6358 kb)
Molecular dynamics simulation of HIV-1 CA pentamer of hexamers
Molecular dynamics simulation of the transition from a relatively flat conformation of a pentamer of hexamers (POH), to a dome-like shape conformation during 300ns. The central pentamer is shown in orange, the surrounding five hexamers are shown in gray. Residues Tyr 184 and Met 185 are shown in spherical representation. (MOV 22123 kb)
Fully equilibrated HIV-1 CA pentamer of hexamers model
Equilibrated structure of the pentamer of hexamers model (POH) in surface representation after 400ns molecular dynamics simulation. The CTDs from both pentamers and hexamers are shown in orange; NTDs belonging to the hexamers are colored in blue, while its pentamer counterpart is shown in green. (MOV 2776 kb)
Cryo-electron tomographic reconstruction of a native HIV-1 core
Computational slices through the 3D tomographic volume of a native HIV-1 core. (MOV 314 kb)
Rights and permissions
About this article
Cite this article
Zhao, G., Perilla, J., Yufenyuy, E. et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013). https://doi.org/10.1038/nature12162
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12162
This article is cited by
-
Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization
Nature Communications (2023)
-
Poxvirus under the eyes of electron microscope
Applied Microscopy (2022)
-
A click chemistry amplified nanopore assay for ultrasensitive quantification of HIV-1 p24 antigen in clinical samples
Nature Communications (2022)
-
Coarse-grained molecular dynamics integrated with convolutional neural network for comparing shapes of temperature sensitive bottlebrushes
npj Computational Materials (2022)
-
A self-complementary macrocycle by a dual interaction system
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.