Abstract
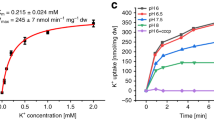
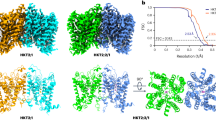
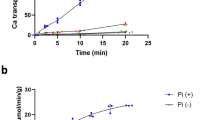
Phosphate is crucial for structural and metabolic needs, including nucleotide and lipid synthesis, signalling and chemical energy storage. Proton-coupled transporters of the major facilitator superfamily (MFS) are essential for phosphate uptake in plants and fungi, and also have a function in sensing external phosphate levels as transceptors1,2,3,4,5. Here we report the 2.9 Å structure of a fungal (Piriformospora indica) high-affinity phosphate transporter, PiPT, in an inward-facing occluded state, with bound phosphate visible in the membrane-buried binding site. The structure indicates both proton and phosphate exit pathways and suggests a modified asymmetrical ‘rocker-switch’ mechanism of phosphate transport. PiPT is related to several human transporter families, most notably the organic cation and anion transporters of the solute carrier family (SLC22), which are implicated in cancer-drug resistance6,7. We modelled representative cation and anion SLC22 transporters based on the PiPT structure to surmise the structural basis for substrate binding and charge selectivity in this important family. The PiPT structure demonstrates and expands on principles of substrate transport by the MFS transporters and illuminates principles of phosphate uptake in particular.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Pao, S. S., Paulsen, I. T. & Saier, M. H., Jr Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34 (1998)
Persson, B. L. et al. Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43, 225–244 (2003)
Nussaume, L. et al. Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2, 83 (2011)
Giots, F., Donaton, M. C. V. & Thevelein, J. M. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47, 1163–1181 (2003)
Popova, Y., Thayumanavan, P., Lonati, E., Agrochao, M. & Thevelein, J. Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc. Natl Acad. Sci. USA 107, 2890–2895 (2010)
Giacomini, K. M. et al. Membrane transporters in drug development. Nature Rev. Drug Discov. 9, 215–236 (2010)
Hediger, M. A. et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch. 447, 465–468 (2004)
Hirai, T. et al. Three-dimensional structure of a bacterial oxalate transporter. Nature Struct. Biol. 9, 597–600 (2002)
Abramson, J. et al. Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615 (2003)
Huang, Y., Lemieux, M. J., Song, J., Auer, M. & Wang, D. N. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301, 616–620 (2003)
Yin, Y., He, X., Szewczyk, P., Nguyen, T. & Chang, G. Structure of the multidrug transporter EmrD from Escherichia coli. Science 312, 741–744 (2006)
Dang, S. et al. Structure of a fucose transporter in an outward-open conformation. Nature 467, 734–738 (2010)
Newstead, S. et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 30, 417–426 (2011)
Solcan, N. et al. Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J. 31, 3411–3421 (2012)
Sun, L. et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490, 361–366 (2012)
Holyoake, J. & Sansom, M. S. P. Conformational change in an MFS protein: MD simulations of LacY. Structure 15, 873–884 (2007)
Smirnova, I. et al. Sugar binding induces an outward facing conformation of LacY. Proc. Natl Acad. Sci. USA 104, 16504–16509 (2007)
Madej, M. G., Soro, S. N. & Kaback, H. R. Apo-intermediate in the transport cycle of lactose permease (LacY). Proc. Natl Acad. Sci. USA 109, 2970–2978 (2012)
Varma, A., Bakshi, M., Lou, B., Hartmann, A. & Oelmueller, R. Piriformospora indica: A novel plant growth-promoting mycorrhizal fungus. Agric. Res. 1, 117–131 (2012)
Yadav, V. et al. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J. Biol. Chem. 285, 26532–26544 (2010)
Fredriksson, R., Nordström, K. J. V., Stephansson, O., Hägglund, M. G. A. & Schiöth, H. B. The solute carrier (SLC) complement of the human genome: phylogenetic classification reveals four major families. FEBS Lett. 582, 3811–3816 (2008)
Schlessinger, A. et al. Comparison of human solute carriers. Protein Sci. 19, 412–428 (2010)
Forrest, L. R., Krämer, R. & Ziegler, C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 1807, 167–188 (2011)
Samyn, D. R. et al. Mutational analysis of putative phosphate- and proton-binding sites in the Saccharomyces cerevisiae Pho84 phosphate:H+ transceptor and its effect on signalling to the PKA and PHO pathways. Biochem. J. 445, 413–422 (2012)
Kaback, H. R., Smirnova, I., Kasho, V., Nie, Y. & Zhou, Y. The alternating access transport mechanism in LacY. J. Membr. Biol. 239, 85–93 (2011)
Krupka, R. M. Coupling mechanisms in active transport. Biochim. Biophys. Acta 1183, 105–113 (1993)
Luecke, H. & Quiocho, F. A. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature 347, 402–406 (1990)
Vyas, N. K., Vyas, M. N. & Quiocho, F. A. Crystal structure of M tuberculosis ABC phosphate transport receptor: specificity and charge compensation dominated by ion-dipole interactions. Structure 11, 765–774 (2003)
Morales, R. et al. Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein. Structure 14, 601–609 (2006)
Feng, B., Dresser, M. J., Shu, Y., Johns, S. J. & Giacomini, K. M. Arginine 454 and lysine 370 are essential for the anion specificity of the organic anion transporter, rOAT3. Biochemistry 40, 5511–5520 (2001)
Mumberg, D., Müller, R. & Funk, M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767–5768 (1994)
Li, M. et al. Selecting optimum eukaryotic integral membrane proteins for structure determination by rapid expression and solubilization screening. J. Mol. Biol. 385, 820–830 (2009)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Pedersen, B. P., Morth, J. P. & Nissen, P. Structure determination using poorly diffracting membrane protein crystals—Lessons from the H+ and Na+,K+-ATPases. Acta Crystallogr. D 66, 309–313 (2010)
Sheldrick, G. M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 (2010)
Terwisscha van Scheltinga, A. C., Valegård, K., Hajdu, J. & Andersson, I. MIR phasing using merohedrally twinned crystals. Acta Crystallogr. D 59, 2017–2022 (2003)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Cowtan, K. ‘dm’: An automated procedure for phase improvement by density modification. CCP4 Newsl. Protein Crystallogr. 31, 34–38 (1994)
Keller, S., Pojer, F., Heide, L. & Lawson, D. M. Molecular replacement in the ‘twilight zone’: structure determination of the non-haem iron oxygenase NovR from Streptomyces spheroides through repeated density modification of a poor molecular-replacement solution. Acta Crystallogr. D 62, 1564–1570 (2006)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
McCoy, A. J. Liking likelihood. Acta Crystallogr. D 60, 2169–2183 (2004)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Petřek, M., Košinová, P., Koča, J. & Otyepka, M. MOLE: A Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure 15, 1357–1363 (2007)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
The PyMOL Molecular Graphics System, Version 1.5.0.4 (Schrödinger LLC, 2012)
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
Pei, J., Kim, B.-H. & Grishin, N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008)
Käll, L., Krogh, A. & Sonnhammer, E. L. L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 (2004)
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Shen, M.-Y. & Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 (2006)
Krivov, G. G., Shapovalov, M. V. & Dunbrack, R. L., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 (2009)
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008)
Bjelkmar, P., Larsson, P., Cuendet, M. A., Hess, B. & Lindahl, E. Implementation of the CHARMM Force Field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 6, 459–466 (2010)
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983)
Vanommeslaeghe, K. et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010)
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012)
Jo, S., Lim, J. B., Klauda, J. B. & Im, W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009)
Berendsen, H. J. C., Postma, J. P. M., Vangunsteren, W. F., Dinola, A. & Haak, J. R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984)
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007)
Parrinello, M. & Rahman, A. Polymorphic transitions in single-crystals - a new molecular-dynamics method. J. Appl. Phys. 52, 7182–7190 (1981)
Acknowledgements
We thank K. Giacomini for discussions about SLC transporters; P. Nissen for comments that improved the manuscript; J. Holton, G. Meigs, C. Ogata and N. Venugopalan for assistance with synchrotron data collection at the Advanced Light Source and Advanced Photon Source; and C. Waddling, P. Wassam and M. Tessema for technical assistance. B.P.P. was supported by a postdoctoral fellowship from the Carlsberg Foundation and later by a fellowship from the Danish Cancer Society; H.K. by a fellowship from Woods Whelan foundation, Council of Scientific and Industrial Research, Government of India, and a travel grant from Jawaharlal Nehru University, New Delhi.; A.Sc. by NIH postdoctoral fellowship F32 GM088991; A.Sa. by NIH grants U54 GM094625 and U01 GM61390; A.K.J. by a Research Assistant Professorship to do work at UCSF from the American Society for Microbiology; and R.M.S. by NIH grants U54 GM094625, GM24485 and GM073210.
Author information
Authors and Affiliations
Contributions
B.P.P. did expression, purification and crystallization experiments, collected and processed the data, and determined, refined and analysed the structure. H.K. identified the target, did purification and crystallization experiments, collected data and identified the use of NG for crystallization. A.B.W. optimized the yeast expression system, and assisted in data collection and data analysis. A.J.R. helped with protein purification and crystallization. Z.R.-Z. assisted in optimization of the yeast expression system, cloned, expressed, purified and characterized the target, and set up initial crystallization experiments. B.H.C. did cloning, expression tests and cell growth. A.Sc. performed bioinformatics analysis and built human homology models. M.B. did molecular dynamics and analysed the results. W.H. trained H.K. and assisted H.K. in data collection. A.Sa. supervised homology modelling, bioinformatics analysis and molecular dynamics. A.K.J. identified the target and initiated the project. R.M.S. supervised the project and analysed the structure. B.P.P. and R.M.S. wrote the paper with input from H.K., A.B.W., A.Sc., A.Sa. and A.K.J.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10, Supplementary Tables 1-2, Supplementary Alignments 1-2 and additional references. (PDF 14710 kb)
Rights and permissions
About this article
Cite this article
Pedersen, B., Kumar, H., Waight, A. et al. Crystal structure of a eukaryotic phosphate transporter. Nature 496, 533–536 (2013). https://doi.org/10.1038/nature12042
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12042
This article is cited by
-
Growth promoting effects of endophytic fungus Piriformospora indica in small cardamom (Elettaria cardamomum Maton)
Symbiosis (2023)
-
Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis
Nature Structural & Molecular Biology (2020)
-
Acanthamoeba castellanii phosphate transporter (AcPHS) is important to maintain inorganic phosphate influx and is related to trophozoite metabolic processes
Journal of Bioenergetics and Biomembranes (2020)
-
Water Availability in Soil Affect Performance of Different Root Fungal Colonizers on Metabolism of Wheat
Iranian Journal of Science and Technology, Transactions A: Science (2020)
-
Expression analysis and functional characterization of two PHT1 family phosphate transporters in ryegrass
Planta (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.