Abstract
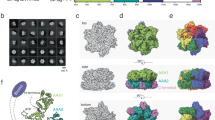
Histone chaperones represent a structurally and functionally diverse family of histone-binding proteins that prevent promiscuous interactions of histones before their assembly into chromatin. DAXX is a metazoan histone chaperone specific to the evolutionarily conserved histone variant H3.3. Here we report the crystal structures of the DAXX histone-binding domain with a histone H3.3–H4 dimer, including mutants within DAXX and H3.3, together with in vitro and in vivo functional studies that elucidate the principles underlying H3.3 recognition specificity. Occupying 40% of the histone surface-accessible area, DAXX wraps around the H3.3–H4 dimer, with complex formation accompanied by structural transitions in the H3.3–H4 histone fold. DAXX uses an extended α-helical conformation to compete with major inter-histone, DNA and ASF1 interaction sites. Our structural studies identify recognition elements that read out H3.3-specific residues, and functional studies address the contributions of Gly 90 in H3.3 and Glu 225 in DAXX to chaperone-mediated H3.3 variant recognition specificity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Atomic structures of the DAXX–H3.3–H4 complex have been deposited in the RCSB Protein Data Bank with accession codes 4H9N (five-substituent native complex at 1.95A˚ ), 4H9S (seven-substituent native complex at 2.60A˚ ), 4H9O (five-substituent H3.3(G90M) mutant complex at 2.05A˚ ), 4H9P (five-substituent H3.3(G90A) mutant complex at 2.20A˚ ), 4H9Q (five-substituent DAXX(E225A) mutant complex at 1.95A˚ ) and 4H9R (five-substituent DAXX(E225A)–H3.3(G90A) mutant complex at 2.20A˚ ). Supplementary Video 1 shows the ternary five-substituent native complex of DAXX–H3.3–H4.
Change history
21 November 2012
A change was made to the title.
References
Hondele, M. & Ladurner, A. G. The chaperon-histone partnership: for the greater good of histone traffic and chromatin plasticity. Curr. Opin. Struct. Biol. 21, 698–708 (2011)
Das, C., Tyler, J. K. & Churchill, M. A. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 35, 476–489 (2010)
Hamiche, A. & Shuaib, M. Chaperoning the histone H3 family. Biochim. Biophys. Acta 1819, 230–237 (2012)
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010)
Wong, L. H. et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 (2010)
Drane, P. et al. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010)
Lewis, P. W. et al. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA 107, 14075–14080 (2010)
Jiao, Y. et al. DAXX/ATRX, MEN1 and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1223 (2011)
Heaphy, C. M. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011)
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodeling genes in pediatric glioblastoma. Nature 482, 226–231 (2012)
Elsässer, S. J. & D’Arcy, S. Towards a mechanism for histone chaperones. Biochim. Biophys. Acta 1819, 211–221 (2012)
Black, B. E. et al. Structural determinants for generating centromeric chromatin. Nature 430, 578–582 (2004)
Sekulic, N., Bassett, E. A., Rodgers, D. J. & Black, B. E. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467, 347–351 (2010)
Hu, H. et al. Structure of CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906 (2011)
Cho, U.-S. & Harrison, S. C. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl Acad. Sci. USA 108, 9367–9371 (2011)
Zhou, Z. et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234–237 (2011)
Luger, K. et al. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997)
Natsume, R. et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 (2007)
English, C. M. et al. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006)
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004)
Levy, Y. & Onuchic, J. N. Water mediation in protein folding and molecular recognition. Annu. Rev. Biophys. Biomol. Struct. 35, 389–415 (2006)
Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. BALBES: a molecular replacement pipeline. Acta Crystallogr. D 64, 125–132 (2008)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Groth, A. et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17, 301–311 (2005)
Das, C., Lucia, M. S., Hansen, K. C. & Tyler, J. K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009)
Acknowledgements
We thank the personnel of synchrotron beam lines 24-I/D-C/E at the Advanced Photon Source (Argonne National Laboratory) and beam line X29 at the Brookhaven National Laboratory for their assistance, and the Center for Synchrotron Biosciences grant, P30-EB-009998, from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) for funding. The use of the Rigaku/MSC microMax 007HF and Formulator in the Rockefeller University Structural Biology Resource Center was made possible by grant numbers 1S10RR022321-01 and 1S10RR027037-01 from the National Center for Research Resources of the NIH. We thank B. Black for sharing data before publication, A. Ruthenburg, J. Song and Z. Cheng for advice and discussions, and E. Datan for help in expressing DAXX protein. D.J.P. was supported by funds from the Abby Rockefeller Mauze Trust, and the Maloris and STARR Foundations. C.D.A. also acknowledges support from the STARR Foundation and The Rockefeller University. J.W.C. acknowledges support from UK Medical Research Council (MRC) (grants U105181009 and UD99999908). S.J.E. was supported by a Boehringer Ingelheim Funds fellowship and the David Rockefeller Graduate Program and holds an EMBO ALTF 1232-2011.
Author information
Authors and Affiliations
Contributions
S.J.E. conceived and led the use of rigidifying mutants to crystallize the complex. Crystals of the seven- and five-substituent complexes were grown by S.J.E. and H.H., respectively. H.H. solved all the crystal structures of the complexes, including mutant complexes, under the supervision of D.J.P. S.J.E. performed biochemical and most cell-based experiments under the supervision of C.D.A and J.W.C. P.W.L carried out cell-based experiments and contributed reagents. All authors discussed the results and commented on the manuscript during its preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18 and Supplementary Tables 1 and 2. (PDF 17204 kb)
Video of DAXX-H3.3-H4 complex
Video of the ternary complex of DAXX (ribbon representation in magenta), H3.3 and H4 (surface representations in blue and green, respectively). The DAXX chaperone in an all alpha-helical conformation envelops the histone H3.3-H4 dimer for H3.3-specific recognition. (AVI 6541 kb)
Rights and permissions
About this article
Cite this article
Elsässer, S., Huang, H., Lewis, P. et al. DAXX envelops a histone H3.3–H4 dimer for H3.3-specific recognition. Nature 491, 560–565 (2012). https://doi.org/10.1038/nature11608
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11608
This article is cited by
-
HIRA vs. DAXX: the two axes shaping the histone H3.3 landscape
Experimental & Molecular Medicine (2024)
-
The histone chaperone function of Daxx is dispensable for embryonic development
Cell Death & Disease (2023)
-
Histone 3.3-related chromatinopathy: missense variants throughout H3-3A and H3-3B cause a range of functional consequences across species
Human Genetics (2023)
-
DAXX drives de novo lipogenesis and contributes to tumorigenesis
Nature Communications (2023)
-
Mechanisms of chromatin-based epigenetic inheritance
Science China Life Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.