Abstract
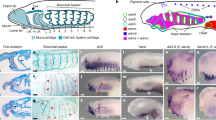
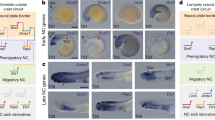
Neuroectodermal signalling centres induce and pattern many novel vertebrate brain structures but are absent, or divergent, in invertebrate chordates. This has led to the idea that signalling-centre genetic programs were first assembled in stem vertebrates and potentially drove morphological innovations of the brain. However, this scenario presumes that extant cephalochordates accurately represent ancestral chordate characters, which has not been tested using close chordate outgroups. Here we report that genetic programs homologous to three vertebrate signalling centres—the anterior neural ridge, zona limitans intrathalamica and isthmic organizer—are present in the hemichordate Saccoglossus kowalevskii. Fgf8/17/18 (a single gene homologous to vertebrate Fgf8, Fgf17 and Fgf18), sfrp1/5, hh and wnt1 are expressed in vertebrate-like arrangements in hemichordate ectoderm, and homologous genetic mechanisms regulate ectodermal patterning in both animals. We propose that these genetic programs were components of an unexpectedly complex, ancient genetic regulatory scaffold for deuterostome body patterning that degenerated in amphioxus and ascidians, but was retained to pattern divergent structures in hemichordates and vertebrates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
S. kowalevskii gene sequences have been deposited in GenBank, and accession numbers are provided in Supplementary Table 2.
References
Echevarria, D., Vieira, C., Gimeno, L. & Martinez, S. Neuroepithelial secondary organizers and cell fate specification in the developing brain. Brain Res. Brain Res. Rev. 43, 179–191 (2003)
Wilson, S. W. & Houart, C. Early steps in the development of the forebrain. Dev. Cell 6, 167–181 (2004)
Wurst, W. & Bally-Cuif, L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nature Rev. Neurosci. 2, 99–108 (2001)
Wicht, H. & Lacalli, T. C. The nervous system of amphioxus: structure, development, and evolutionary significance. Can. J. Zool. 150, 122–150 (2005)
Lacalli, T. C. Prospective protochordate homologs of vertebrate midbrain and MHB, with some thoughts on MHB origins. Int. J. Biol. Sci. 2, 104–109 (2006)
Meinertzhagen, I. A., Lemaire, P. & Okamura, Y. The neurobiology of the ascidian tadpole larva: recent developments in an ancient chordate. Annu. Rev. Neurosci. 27, 453–485 (2004)
Lowe, C. J. et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 113, 853–865 (2003)
Holland, L. Z. & Short, S. Gene duplication, co-option and recruitment during the origin of the vertebrate brain from the invertebrate chordate brain. Brain Behav. Evol. 72, (2008)
Holland, L. Z. Chordate roots of the vertebrate nervous system: expanding the molecular toolkit. Nature Rev. Neurosci. 10, 736–746 (2009)
Irimia, M. et al. Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. Evodevo. 1, 7 (2010)
Tomer, R., Denes, A. S., Tessmar-Raible, K. & Arendt, D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142, 800–809 (2010)
Urbach, R. A procephalic territory in Drosophila exhibiting similarities and dissimilarities compared to the vertebrate midbrain/hindbrain boundary region. Neural Dev. 2, 23 (2007)
Crossley, P. H., Martinez, S. & Martin, G. R. Midbrain development induced by FGF8 in the chick embryo. Nature 380, 66–68 (1996)
Reifers, F. et al. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125, 2381–2395 (1998)
Houart, C. et al. Establishment of the telencephalon during gastrulation by local antagonism of Wnt signaling. Neuron 35, 255–265 (2002)
Paek, H., Gutin, G. & Hebert, J. M. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development 136, 2457–2465 (2009)
Kiecker, C. & Lumsden, A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nature Neurosci. 7, 1242–1249 (2004)
Scholpp, S., Wolf, O., Brand, M. & Lumsden, A. Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development 133, 855–864 (2006)
Meulemans, D. & Bronner-Fraser, M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS ONE 2, e787 (2007)
Imai, K. S., Stolfi, A., Levine, M. & Satou, Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136, 285–293 (2009)
Shimeld, S. M. The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Dev. Genes Evol. 209, 40–47 (1999)
Takatori, N., Satou, Y. & Satoh, N. Expression of hedgehog genes in Ciona intestinalis embryos. Mech. Dev. 116, 235–238 (2002)
Scholpp, S. & Lumsden, A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 33, 373–380 (2010)
Bertrand, S. et al. Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc. Natl Acad. Sci. USA 108, 9160–9165 (2011)
Holland, L. Z., Holland, N. N. & Schubert, M. Developmental expression of AmphiWnt1, an amphioxus gene in the Wnt1/wingless subfamily. Dev. Genes Evol. 210, 522–524 (2000)
Bourlat, S. J. et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature 444, 85–88 (2006)
Darras, S., Gerhart, J., Terasaki, M., Kirschner, M. & Lowe, C. J. β-catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii. Development 138, 959–970 (2011)
Gillis, J. A., Fritzenwanker, J. H. & Lowe, C. J. A stem-deuterostome origin of the vertebrate pharyngeal transcriptional network. Proc. R. Soc. B 279, 237–246 (2012)
Lowe, C. J. et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 4, e291 (2006)
Shimamura, K. & Rubenstein, J. L. Inductive interactions direct early regionalization of the mouse forebrain. Development 124, 2709–2718 (1997)
Fukuchi-Shimogori, T. & Grove, E. A. Neocortex patterning by the secreted signaling molecule FGF8. Science 294, 1071–1074 (2001)
Walshe, J. & Mason, I. Unique and combinatorial functions of Fgf3 and Fgf8 during zebrafish forebrain development. Development 130, 4337–4349 (2003)
Garel, S., Huffman, K. J. & Rubenstein, J. L. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development 130, 1903–1914 (2003)
Mohammadi, M. et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955–960 (1997)
Lagutin, O. V. et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 17, 368–379 (2003)
Crossley, P. H., Martinez, S., Ohkubo, Y. & Rubenstein, J. L. Coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon during development of the telencephalic and optic vesicles. Neuroscience 108, 183–206 (2001)
Hébert, J. M. & Fishell, G. The genetics of early telencephalon patterning: some assembly required. Nature Rev. Neurosci. 9, 678–685 (2008)
Zeltser, L. M., Larsen, C. W. & Lumsden, A. A new developmental compartment in the forebrain regulated by Lunatic fringe. Nature Neurosci. 4, 683–684 (2001)
Scholpp, S. et al. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development 134, 3167–3176 (2007)
Chen, J. K., Taipale, J., Cooper, M. K. & Beachy, P. A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 (2002)
Simon, H. H., Thuret, S. & &Alberi, L. Midbrain dopaminergic neurons: control of their cell fate by the engrailed transcription factors. Cell Tissue Res. 318, 53–61 (2004)
McMahon, A. P., Joyner, A. L., Bradley, A. & McMahon, J. A. The midbrain-hindbrain phenotype of Wnt-1−Wnt-1− mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69, 581–595 (1992)
Nomaksteinsky, M. et al. Centralization of the deuterostome nervous system predates chordates. Curr. Biol. 19, 1264–1269 (2009)
Gavino, M. A., Reddien, P. W. & A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr. biol. 21, 294–299 (2011)
Grande, C. & Patel, N. H. Nodal signalling is involved in left–right asymmetry in snails. Nature 457, 1007–1011 (2009)
Campo-Paysaa, F., Marletaz, F., Laudet, V. & Schubert, M. Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis 46, 640–656 (2008)
Freeman, R. M., Jr et al. cDNA sequences for transcription factors and signaling proteins of the hemichordate Saccoglossus kowalevskii: efficacy of the expressed sequence tag (EST) approach for evolutionary and developmental studies of a new organism. Biol. Bull. 214, 284–302 (2008)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003)
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001)
Acknowledgements
We thank J. Gerhart and M. Kirschner for assistance and support, R. Freeman for bioinformatics assistance, E. Farrelly, M. Terasaki and S. Darras for technical guidance, the members of the Lowe laboratory for discussions, and G. Wray and J. Gerhart for comments on drafts of the manuscript. We also thank the staff of the Marine Biological Laboratory, the Waquoit Bay National Estuarine Research Reserve, Carl Zeiss and Nikon for assistance during our field season. This work was funded by the Searle Kinship Foundation, Brain Research Foundation and National Science Foundation grant 1049106 (C.J.L.), National Institutes of Health grant R01 HD42330 (E.A.G.) and the University of Chicago Hinds Fund (A.M.P). A.M.P. was supported by a Marine Biological Laboratory Frank R. Lillie Fellowship, National Institute of Child Health and Development institutional training grant 1T32HD055164-01A1, and National Institute of Neurological Disorders and Stroke pre-doctoral fellowship 1F31NS074738-01A1. J.A. was supported by a National Science and Engineering Research Council of Canada pre-doctoral grant.
Author information
Authors and Affiliations
Contributions
A.M.P., C.J.L. and J.A. conceived the project. A.M.P. and C.J.L. performed the hemichordate experiments and wrote the paper. E.E.M. and S.A. performed mouse experiments, and E.A.G. edited the paper. All authors discussed and commented on the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-4 and Supplementary Tables 1-2. Please note that Supplementary Figure 1 shows that fz5/8 siRNA affects proboscis patterning specifically and that Supplementary Figure 2 shows Ptch expression in wild-type S. kowalevskii embryos and spectrum of phenotypes after hh siRNA injection. Ptch expression indicates that hh can signal to numerous body regions. Hh siRNA injection causes pleiotropic effects on AP and DV patterning. (PDF 2369 kb)
Rights and permissions
About this article
Cite this article
Pani, A., Mullarkey, E., Aronowicz, J. et al. Ancient deuterostome origins of vertebrate brain signalling centres. Nature 483, 289–294 (2012). https://doi.org/10.1038/nature10838
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10838
This article is cited by
-
The Cambrian fossil Pikaia, and the origin of chordate somites
EvoDevo (2024)
-
Three topological features of regulatory networks control life-essential and specialized subsystems
Scientific Reports (2021)
-
GABAergic motor neurons bias locomotor decision-making in C. elegans
Nature Communications (2020)
-
Step-wise evolution of neural patterning by Hedgehog signalling in chordates
Nature Ecology & Evolution (2020)
-
Gene expression analysis of three homeobox genes throughout early and late development of a feather star Anneissia japonica
Development Genes and Evolution (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.