Abstract
The development of high throughput technologies based on the knowledge of the human genome has opened the possibility to search for global genomic alterations in tumors responsible for their development and progression that may have important clinical implications. One of the major applications of this genomic knowledge has been the design of different types of microarray platforms for the analysis of DNA alterations and gene expression profiling (GEP). The main contributions of the DNA studies in lymphoid neoplasms include the definition of relatively characteristic genomic profiles for specific disease entities, the demonstration of common chromosomal alterations across entities, the identification of genes and pathways targeted by the altered chromosomal regions, and the identification of chromosomal alterations with prognostic implications. RNA GEP studies in these tumors have enhanced the molecular characterization of known entities and facilitated the recognition of new subtypes and categories of lymphoid neoplasms, the identification of new biomarkers and prognostic models, and the detection of oncogenic pathways with potential implications for targeted therapies. The recent development of the next generation sequencing (NGS) technologies and its application in lymphoid neoplasms already have provided an initial view of the complex landscape of somatic mutations in these tumors and some findings with important functional and clinical implications. This review addresses the major contributions and limitations of the microarray technologies in the understanding of lymphoid neoplasms and discusses how this knowledge may be transferred into the clinics. The initial results of the NGS studies are also presented.
Similar content being viewed by others
Main
Hematopathology has advanced in parallel with technological developments that have expanded our understanding of the phenotypic, genetic, and molecular characteristics of the hematological neoplasms. The translation of this knowledge into clinical practice has changed the conceptual framework of our work over the years with an increased ability to generate more precise and reproducible diagnoses of the different entities and sustaining the progressive expansion of more effective and tailored therapeutic strategies.
The initial elucidation of the Human Genome a decade ago and the development of high throughput technologies opened the possibility to search for comprehensive views of the genomic alterations responsible for tumor development and progression (Figure 1). One of the major applications of this genomic knowledge in the study of lymphoid neoplasms has been the design of different types of microarray platforms for the global analysis of DNA alterations and gene expression profiling (GEP).1, 2 The recent development of a new generation of sequencing technologies (next generation sequencing (NGS) or massively parallel sequencing) and their systematic application to human cancer and in particular to lymphoid neoplasms are revealing a landscape of somatic mutations of unprecedented complexity.3 These studies already have provided a number of important findings with functional and clinical implications including new potential targets for advanced therapies. It is perhaps too early to define what will be the real influence of the new knowledge gained from NGS in clinical practice but the perspectives are challenging and promising. This review addresses the major contributions and limitations of the microarray technologies in diagnostic hematopathology and the initial results of the NGS studies in lymphoid neoplasms that will give us a glimpse of what these studies may offer us in the very near future.
Microarray technologies
The use of DNA microarrays has represented a major technological advance in the study of lymphoid neoplasms. The information generated has had an important impact in the understanding of their genetic and molecular pathogenesis that has supported changes in the recognition and diagnosis of some tumor subtypes and different lymphoma categories. However, it has been difficult to translate some of these findings into clinical practice.
Different platforms have been developed based on DNA fragments cloned in different vectors or on oligonucleotides of short or longer length, which require slightly different technical approaches but the information generated is relatively similar and robust. These platforms have been designed for the study of RNA GEP and DNA changes, including chromosomal copy number alterations (CNA), genotyping, and epigenetic modifications. Integrative studies using both types of platforms for DNA and RNA have facilitated the discovery of target genes and pathways that, together with global signatures and genomic profiles, have provided new perspectives in the understanding of lymphoid neoplasms.
Microarrays for DNA studies
Platforms
The first arrays for DNA studies followed the strategy introduced by the comparative genomic hybridization (CGH) technique, which allows the detection of unbalanced DNA copy number changes. In this approach, tumor and a reference normal DNA of the same gender are labeled with different fluorochromes and competitively hybridized on normal chromosome metaphase spreads. The different intensity of the tumor and normal DNA hybridization signal indicates the presence of gains or losses in specific chromosomal regions of the tumor. In the initial CGH arrays, the metaphase spreads were substituted by DNA fragments cloned in plasmid, bacterial, or yeast artificial chromosomes, and more recently by long oligonucleotides (50–75-mer).1 The current high density oligonucleotide arrays cover the whole genome (WG) with probes spaced about 1–5 kb apart. The single-nucleotide polymorphism arrays (SNP arrays) are alternative platforms to the CGH arrays. These arrays use short oligonucleotides (25-mer) that distinguish the different genotypes of a given SNP. The distribution of the probes throughout the genome is variable and depends on the number of known SNPs with some areas of the genome less covered than others. The more recent SNP arrays also contain probes to identify regions of copy number variants (CNV), a major source of individual genetic variation (Figure 2). CNV are DNA segments of 1 kb or larger that are present in constitutional DNA at variable copy number in comparison with a reference genome. They may have a simple structure, such as tandem duplication of a single copy, or may involve complex gains, losses, or inversions of homologous sequences.4
Germline copy number variants (CNV) and somatic copy number alterations. (a) CNV are DNA segments present in constitutional DNA at variable copy number in comparison with a reference genome. They may have different structures. (b) In the genomic analysis of a tumor DNA, CNV can be misinterpreted as copy number alterations (gains or amplifications in the figure) if the tumor DNA is not compared with a reference normal DNA or compared with non-related constitutional DNA from other individuals. However, the use of the constitutional DNA of the same patient clearly identifies these regions as CNV already present in the normal DNA and identifies a region of acquired deletion.
The two types of high density platforms have advantages and disadvantages and provide complementary views of the genome (Table 1). Both CGH and SNP arrays allow the detection of CNA at high resolution but both require approximately at least 30% of cells carrying the same abnormal region to be detected.1 This sensitivity may turn into an advantage since it may filter out small irrelevant clones, although, on the other hand, it may miss small clones that may be important later on in the evolution of the disease. These platforms also allow the use of DNA extracted from routinely processed formalin-fixed and paraffin-embedded (FFPE) tissues.5 CGH arrays have an increased sensitivity, have a more homogeneous coverage of the genome and may detect small alterations with higher resolution but they need the combination of a normal reference DNA with the tumor DNA. The use of the constitutional DNA of the same patient provides the advantage of filtering out the individual CNV. These regions may be interpreted as gains or losses if the tumor DNA is compared with non-related constitutional DNA from other individuals. Although high resolution maps of CNV are available to interpret the results, the use of reference DNA from the same individual facilitates the analysis (Figure 2).6 The SNP arrays provide genotype information that has also been used for genome-wide association studies to determine the relationship between single SNPs and the risk for specific diseases including lymphoid neoplasms.7
One of the major advantages of the SNP arrays is the possibility to detect regions of Uniparental Disomy (UPD) or DNA copy-neutral loss of heterozygosity in addition to the DNA copy number changes.8 These regions correspond to stretches of DNA in which both strands are identical and therefore all the SNP are homozygous. The genotyping information of the SNP arrays recognizes these regions. The simultaneous measurement of the copy number based on the intensity of the hybridization signal identifies the presence of two strands of DNA in these regions and distinguishes them from the homozygosity generated by a deletion of the chromosomal region in which the hybridization signal would be weaker corresponding to only one strand of DNA. The use of SNP arrays has shown that UPD is more frequent than initially thought both in constitutional and in tumor DNA and they may be relevant in the pathogenesis of the neoplasms. UPD in tumors may occur by different mechanisms but usually implies the deletion of one allele and the correction of the defect by the duplication of the remaining allele. In this way, UPD may reduce to homozygosity a mutated allele after the deletion of the normal allele and the duplication of the mutated one. This phenomenon has been observed in mutations of TP53 in which both alleles carry the same mutation but also associated with activating mutations such as JAK2 mutations in myeloproliferative neoplasms.8, 9
Contributions of DNA Microarrays in Lymphoid Neoplasms
DNA-array studies have expanded and refined the initial findings obtained with the metaphase CGH. The main contributions may be summarized as follows (Table 1):
-
1
Definition of genomic profiles relatively characteristic of each disease entity as well as common chromosomal alterations across entities;
-
2
Identification of genes and pathways targeted by the altered chromosomal regions;
-
3
Description of chromosomal alterations with prognostic implications.
Several lymphomas are genetically characterized by the presence of an alteration that occurs in virtually all cases. In some of them, it is the sole abnormality detected suggesting that it may be the primary lesion required for the development of the tumor. Most of these alterations are chromosomal translocations targeting specific oncogenes such as BCL2, MYC, CCND1, and others.10 The systematic analysis of chromosomal imbalances using metaphase- and array-CGH has also shown that most lymphomas carry a higher number of secondary chromosomal alterations than previously observed by conventional cytogenetic studies. Although some regions are recurrently found in different lymphomas, the global profile is relatively specific for each disease entity, suggesting that these alterations may have a role in defining the biological behavior of the tumor. In each lymphoma type, the number and distribution of the genomic alterations may vary from patient to patient and this heterogeneity may account, in part, for the different behavior of the tumor among patients with the same disease.11, 12, 13 Examples of this are chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL) that have frequent deletions in 13q and 11q and gains of chromosome 12. However, gains of 3q and losses of 1p are relatively frequent in MCL but not in CLL.14, 15, 16, 17 Gains or trisomy of chromosome 3 are also a frequent feature of marginal zone lymphomas (MZL) and diffuse large B-cell lymphoma (DLBCL) of the activated B-cell subtype (ABC).18, 19 These two types of tumors have frequent gains in 18q but ABC-DLBCL also has frequent deletions in 6q, 9p, and 19q not common in MZL.19 DLBCL of germinal center B-cell type (GCB) and primary mediastinal B-cell lymphoma (PMBL) differ genetically from the ABC subtype. These two subtypes do not have the common alterations of the ABC subtype but carry frequent gains in 2p that are uncommon in the ABC-DLBCL. Gains of 9p are seen in PMBCL but are rare in the other two types of DLBCL. Interstitial losses of 7q are a frequent finding in splenic MZL (SMZL) but less frequent in MCL and rare in CLL or other small B-cell lymphomas.12, 20 Differences in genomic profiles have also been observed in different types of peripheral T (PTCL) and NK-cell lymphomas. PTCL-NOS and angioimmunoblastic T-cell lymphoma (AITL) share gains in chromosome 8, 9, and 19 and losses in chromosome 2 but also vary in different regions.21 ALK-positive and -negative anaplastic large cell lymphomas (ALCL) have different profiles that also differ from those seen in PTCL.22 Enteropathy-associated T-cell lymphoma has frequent gains at 9q33 that not are not seen in other T-cell lymphomas.23, 24
In spite of the marked heterogeneity of the genomic profiles in different lymphoma types, some alterations are recurrently seen across different entities, suggesting that they may deregulate crucial genes or pathways in the biology of the tumors, independent of the cell type. For instance, deletions of 17p targeting TP53 and homozygous deletions of 9p21 including CDKN2A are seen in aggressive forms of different lymphomas.11, 25 The amplicon at 13q31 targeting the miR-17-92 cluster has been observed in Burkitt’s lymphoma (BL), MCL, and ABC-DLBCL.14, 19, 26 Deletions of different regions of 6q are seen in different types of lymphomas but the common involved regions may vary and the relevant genes are not well characterized.11, 14, 19
The delineation of minimal common deleted or amplified chromosomal regions and the integrative analysis with gene expression profiles and functional studies have been useful strategies to identify the target genes of recurrent chromosomal alterations. Thus, MIR17HG (host gene of the miR-17-92 cluster) is the only gene included in the minimal amplified region of 13q31 in MCL and the amplification is associated with overexpression of all the miRs in the cluster.14, 27 The inhibitor of NFκB TNFAIP3/A20 was initially identified as the target of the 6q23 deletions found in ocular MZL by array-CGH.28 Inactivating mutations of this gene were subsequently found in other lymphomas carrying 6q23 deletions including ABC-DLBCL, MCL, and Hodgkin’s lymphoma (HL).28, 29 Inactivating mutations of PRDM1/BLIMP1 occur in DLBCL with deletions of 6q21. A recent combined CGH array and gene expression profiling study of NK-cell neoplasms has also recognized 6q21 deletions and inactivating PRDM1 mutations in these tumors.30
Most recurrently altered chromosomal regions include several genes. A lesson recently learned is that these regions may harbor more than one ‘driver’ gene having a cooperative effect in the biology of the tumor cells. PMBL and HL have recurrent amplification of 9p24. The amplicon includes the kinase JAK2 and the histone demethylase JMJD2C. The simultaneous amplification and overexpression of these genes has an additive effect modulating the expression of several genes including MYC.31 Interestingly, these tumors also have deletions in 16p13 associated with inactivating mutations of SOCS1, a negative regulator of JAK2.32 SOCS1 inactivation by biallelic mutations or mutations and deletions promotes the activity of JAK2. The presence of 9p24 amplification and 16p13 deletions in PMBL shows how different chromosomal aberrations in the same type of tumor target genes in the same pathogenetic pathway. Similar findings have been observed in other tumors. For instance, in MCL recurrent chromosomal alterations individually occurring at low frequency, target multiple genes of the same pathways including cell-cycle regulation, DNA damage response, and cell survival (Figure 3).14 SNP array studies have recently revealed that the deletion of several genes of the Hippo signaling pathway in MCL, a mechanism controlling proliferation and apoptosis, may be involved in lymphomagenesis.9
The presence of recurrent chromosomal alterations in cancer does not always reflect a positive selection for activated or inactivated genes with a ‘driver’ function in the pathogenesis of the tumor. Many chromosomal deletions actually do not include genes but are located at known fragile sites, and thus are just a manifestation of genomic instability of the tumor.33 Local structural features of certain DNA regions may also influence genetic alterations. The regions flanking somatic UPD in MCL are significantly enriched in CNV and segmental duplications, suggesting that these regions may facilitate the recombination of DNA.14
As with conventional cytogenetic and CGH studies, array analyses have identified a number of chromosomal alterations related to patient outcomes. Some of these regions reflect a well-known underlying molecular alteration, such as the inactivation of TP53 or CDKN2A in 17p or 9p21, respectively, with direct implications in the biology of the tumor.15, 19 However, the mechanism related to the poor outcome is not as evident with other regions. For instance, large gains of chromosome 3 have been associated with aggressive behavior in ABC-DLBCL and MCL but the potential target is still not known.15, 19 In some lymphoid neoplasms, such as CLL and MCL, DNA-array studies have shown that the genomic complexity is an important prognostic parameter independent of other known factors.15, 16, 17
Gene expression profiling
The microarray technologies have made possible the study of the global GEP of tumors (Table 2). These platforms consist of numerous DNA probes immobilized on a solid surface. The initial arrays were homemade and usually constructed with cDNA probes but they have been progressively substituted by commercially available platforms that use oligonucleotide probes.34 The RNA of the sample is labeled with a fluorochrome and hybridized on the array. The signal obtained reflects the concentration of the corresponding transcript. The results give a quantitative measure and are highly reproducible using the same array platform and also among different platforms.35 Given the high number of genes measured simultaneously, the evaluation of global gene signatures seems more robust than measuring individual genes.34 As in all genomic studies, major challenges are the bioinformatics tools and methodologies to analyze and validate the data using other technologies such as PCR or immunohistochemistry and in other independent series of samples. The systematic analysis of lymphoid neoplasms has provided relevant information in three major areas:
-
1)
Molecular characterization of known entities and recognition of new subtypes and categories of lymphoid neoplasms.
-
2)
Identification of new biomarkers and prognostic models.
-
3)
Detection of oncogenic pathways with implications for targeted therapies.
Characterization of Known Entities and Recognition of New Categories of Lymphoid Neoplasms
Diffuse large B-cell lymphomas
GEP studies of lymphoid neoplasms have revealed that each major lymphoma entity is characterized by a unique and robust program of gene expression.36, 37 This proof of concept has sustained the discovery of some new categories and tumor subtypes with relevance in clinical practice. One of the major contributions of these studies has been the identification of two major subgroups in the category of DLBCL, the GCB-DLBCL, and the ABC-DLBCL. In addition, the profile of these subtypes is also different from the GEP of PMBL.38, 39, 40 The GCB tumors are characterized by the expression of genes related to germinal center cells, whereas ABC tumors have an expression pattern related to mitogenically activated B cells close to cells with a secretory function.36, 38 ABC and PMBL, but not GCB-DLBCL, have constitutive activation of the NFκB pathway, that they require for survival and therefore, it may be an interesting target for therapy. The activation of this pathway in ABC tumors seems to occur through BCR signaling with acquired activating mutations in elements of this pathway including CD79a, CARD11, and MYD88 and inactivating mutations of the NFκB inhibitor A20.29, 31 The clinical, pathological, and biological features of these two types of molecular DLBCL are different, supporting the idea that they may correspond to different entities (reviewed by J Said in this course).41, 42
Other expression profiling studies have identified alternative subgroups of DLBCL characterized by expression signatures related to potential pathogenetic mechanisms. In particular, one subgroup was characterized by high expression of genes associated with oxidative phosphorylation (OxPhos subgroup) and mitochondrial function such as genes of the BCL2 family. The ‘BCR/Proliferation’ subgroup was enriched for B-cell receptor signaling and cell-cycle regulatory genes whereas the ‘Host Response’ (HR) subgroup had increased expression of genes related to an inflammatory/immune response signature.43 Unlike with the GCB vs ABC categories, these three subgroups were not of prognostic importance. However, they may still be of clinical interest in suggesting possible new therapeutic strategies.
Small B-cell lymphoid neoplasms
The GEP of the most frequent categories of small B-cell neoplasms has provided important insights into the understanding of these diseases and recognized subtypes with clinical implications. CLL and hairy cell leukemia (HCL) have a GEP related to memory B cells with specific features different from other B-cell neoplasms.44, 45, 46 The two major subtypes of CLL with mutated and unmutated IGHV also have some differences in their expression profile. The GEP of follicular lymphoma (FL) has revealed the cell complexity of the microenvironment and its influence in the prognosis, signatures related to the aggressiveness of the tumor or associated with the transformation to DLBCL.47, 48, 49 GEP studies of MCL have recognized a variant of cyclin D1-negative tumors that share the same molecular profile with the conventional cyclin D1-positive cases.37, 50 In addition, non-nodal MCL with a very indolent clinical behavior have a global GEP more similar to conventional MCL than to other small B-cell lymphomas,51 suggesting that they correspond to the same disease. However, these two subgroups also differ in the expression of a number of genes, as well as in other clinical and biological features, suggesting that they may correspond to a specific subtype of the disease.51, 52 The expression program of marginal zone lymphomas shows that nodal and extranodal types share common profiles with upregulation of genes related to cell–cell and cell–extracellular matrix interactions.53, 54
Peripheral T-cell lymphomas
GEP studies in T-cell lymphomas have been able to recognize specific global expression patterns for the most common entities although they are more difficult to interpret, due to the lower frequency of these tumors and the heterogeneity of the tumor cell microenvironment. PTCL, NOS tend to form a cluster in GEP studies but some of these tumors also cluster with other molecular subtypes, indicating the heterogeneity of this category. GEP has identified a possible subgroup of cytotoxic PTCL.55 AITL had a strong signature related to normal follicular T-helper cells and overexpression of genes related to the varied components of the microenvironment including B cells, follicular dendritic cells, extracellular matrix, and angiogenesis.56, 57 ALK-positive and -negative ALCL differ in their GEP and both are different from that of PTCL, NOS, supporting their distinction in the WHO classification.58 NK-cell lymphomas and hepatosplenic lymphomas also have a distinctive GEP. A group of γδ T-cell lymphomas share profiling features with NK-cell lymphomas.59
Burkitt’s lymphoma and B-cell lymphoma unclassifiable with features intermediate between BL and DLBCL
Two studies have described the GEP of BL that refine the differential diagnosis with DLBCL.60 The BL signature shows high expression of MYC targets and genes related to germinal center cells and low expression of NFκB targets and MHC class I genes.60 Intriguingly, although there was a good correlation between the pathology and molecular diagnosis, the discordances were also striking. Some cases expressing the molecular signature of BL (mBL) were diagnosed by expert pathologists as DLBCL or high-grade B-cell lymphomas. On the contrary, only a minority of cases without the molecular signature of BL had been called BL. Curiously, both studies identify occasional cases of molecular BL that lacked a demonstrable MYC rearrangement. Both studies revealed that the molecular distinction between BL and DLBCL in some cases is not very sharp. Tumors with an expression signature intermediate between BL and DLBCL or discordant between the mBL signature and the pathology diagnosis of DLBCL or high-grade lymphoma had frequent expression of BCL2, carried the t(8;14) and frequently additional BCL2 or BCL6 rearrangements (double hit), had more complex karyotypes, presented in older patients and had a worse outcome than cases in which both the molecular and pathology diagnosis were in agreement.60 These observations suggest that some aggressive lymphomas may have molecular and pathological features intermediate between BL and DLBCL and support the proposal of the WHO classification10 recognizing this intermediate category, not as a specific entity but as a biologically heterogeneous category that should be recognized and studied separately.
B-cell lymphoma unclassifiable with features intermediate PMBL and HL
One striking finding of the GEP studies of PMBCL was the similarity with the GEP of HL.39, 40 The major difference was the downregulation of the B-cell differentiation program in HL. These molecular findings further support the previous clinical and pathological observation of a very close relationship between PMBL and particularly mediastinal classical HL (cHL) (reviewed in this issue by Harris).61, 62 Together with more recent genetic and epigenetic studies, they also support the inclusion in the WHO classification of the category large B-cell lymphoma, unclassifiable with features intermediate between DLBCL and HL.63, 64, 65, 66
Identification of New Biomarkers and Prognostic Models
The discovery of clinically significant individual gene expression and gene expression signatures based on GEP studies have also led to more routine tests that have been incorporated into clinical practice for diagnostic or prognostic purposes. ZAP70 expression was found as one of the best discriminatory genes between IGHV-mutated and -unmutated CLL44 and its detection by flow cytometry or immunohistochemistry has been introduced in clinical studies.67, 68 Annexin A1 was identified as a specific marker of HCL that could help in the differential diagnosis with other small B-cell neoplasms with a widely used immunohistochemical stain now commercially available.46 SOX11 overexpression was found as a relatively specific feature of MCL since it was detected in virtually all MCL but absent in other mature B-cell lymphomas with the exception of some BL.69, 70 SOX11 was also expressed in cyclin D1-negative MCL and therefore it is a good biomarker for the diagnosis of this variant.70 SOX11 was also an element of the gene signature distinguishing conventional MCL from a subgroup of MCL with a very indolent clinical behavior, since it was negative in these latter tumors. The use of this marker, together with other clinical (non-nodal presentation) and biological features (absence of genomic complexity and 17p/TP53 alterations), may be useful to recognize this particular subgroup of MCL.51, 52, 71
Microarray studies in malignant lymphomas have provided new and robust prognostic information that improves the current prognostic indices mainly based on clinical criteria, such as the International Prognostic Index (IPI). Interestingly, the GEP-based prognostic models are different for each disease entity, suggesting that the behavior of each lymphoma is determined by different mechanisms. Thus, in DLBCL the best predictors of survival in patients treated with immunochemotherapy include the signatures related to the cell of origin combined with signatures reflecting different cell populations of the tumor microenvironment. The GCB-DLBCL subtype and a signature related to extracellular matrix deposition and inflammatory cell infiltration are associated with better outcome, whereas the ABC-DLBCL subtype and a signature reflecting angiogenesis predict for poorer survival.41 In FL, the best predictor model of survival combines a favorable signature mainly composed of T cell-related genes and an unfavorable profile enriched in macrophage-related genes.47 The GEP studies of MCL confirmed the value of cell proliferation as the best prognostic parameter.37 Proliferation also seems to emerge as one of the best prognostic factors in PTCL.72 These prognostic models, derived from expression profiling analyses, very precisely stratify the patients according to their risk based on quantitative models that also reflect the biology of the disease.
Detection of Oncogenic Pathways with Implications for Targeted Therapies
The oncogenic pathways identified by GEP in different types of tumors may become targets of novel drugs or identify patients susceptible to different therapeutic strategies. Experimental studies have shown that activation of the NFκB pathway is required for survival in ABC-DLBCL and therefore may be an interesting target for therapy.73 The activation of this pathway in PMBL but not in GCB-DLBCL may also suggest a selective indication for target therapy in these tumors. A subset of ABC-DLBCL has activation of the STAT3 pathway, suggesting that this pathway may be a potential therapeutic target in these tumors.74, 75 The molecular classification of DLBCL may be important to select different therapeutic modalities given the apparent difference in their response to certain treatments.76 The high expression of PDFGRα detected in PTCL may be a potential target for therapy in these lymphomas.56 The emerging information in this field and the increasing availability of new drugs designed to target specific molecular genes or pathways emphasizes the need to incorporate this knowledge into clinical practice.77 The use of microarray studies in the context of well-designed clinical trials may have a role in predicting the clinical response to specifically oriented molecular therapies.
How should genomic knowledge be transferred into the clinic?
The application in clinical practice of what has been learned based on the current microarray technology remains an important challenge. The application of individual biomarkers for diagnosis such as ZAP70, Annexin A1, or SOX11 has been incorporated using flow cytometry or routine immunohistochemistry. The incorporation of more global information generated by DNA and GEP arrays is more difficult. This approach requires the extraction of good quality DNA and RNA from tissues or blood. The requirement of fresh samples is a logistic challenge difficult to overcome in routine practice. However, recent improvements in protocols that use nucleic acid extracted from FFPE routinely processed tissues for microarrays increases the likelihood that these technologies can be used in routine practice.5, 78 The information generated up to now has been based on the use of frozen samples. Validation studies using these new protocols for routine samples will be necessary to confirm the applicability of the results.
The information generated with the microarray studies may be translated into the clinics using different platforms such as FISH for genetic studies and quantitative PCR (qPCR), other mRNA detection techniques or immunophenotyping for expression information. These approaches may be useful for small numbers of genes but may not perform well when algorithms using a high number of genes may be needed. Several studies have obtained promising results in the diagnosis of molecular subtypes of DLBCL or applying the MCL proliferation signature using a small number of genes by qPCR or alternative techniques such as RNAse protection assay.79, 80, 81
The most widely used method for translation of GEP contributions into clinical practice is immunohistochemistry; however, there are a variety of difficulties that still need to be overcome. The immunohistochemical detection of the proliferation activity as a prognostic factor in MCL using Ki67 seems robust in many different studies and it has been incorporated into an integrated prognostic model with clinical parameters.82 However, the limitation, as with other immunohistochemical studies, is the reproducibility of the evaluation among pathologists.83 Different algorithms and individual markers have been designed to capture the molecular classification of DLBCL and the prognostic value of different GEP-based prognostic models. The results among different studies are conflicting for varied reasons, probably including case selection and technical difficulties.84 The evaluation of the immunohistochemical results has not overcome the limitations in standardization procedures and evaluation. Pathologists perform well with some markers but the reproducibility in the precise quantification needed in some algorithms is relatively poor.83 A major limitation is the complexity of the GEP signatures that usually incorporate multiple genes, whereas immunohistochemical approaches use a very limited number of markers. On the other hand, GEP models provide a quantitative measure of the expression whereas immunohistochemical results are difficult to quantify. Computerized assisted approaches have been assessed and apparently they may overcome part of the reproducibility and quantitative limitations.85 However, these methods are difficult to establish universally, time consuming and may be difficult to incorporate on a routine basis. The need to transfer the genomic information into clinical practice will increase with the development of new therapies. It is still difficult, however, to foresee which methodologies will allow for this to be accomplished in diagnostic hematopathology. The challenges are overwhelming for individual groups and will require the collaborative efforts of large consortiums and a high dose of imagination.
Next generation sequencing
Principles and Applications
The new generation of sequencing technologies is expanding the possibilities to analyze the mutational spectrum of cancer genomes with a comprehensive perspective thanks to their high speed, relative low cost, and versatility to detect all types of genomic alterations. Several methodologies have been developed that start with the fragmentation of the DNA and subsequent amplification. The multiple fragments generated are simultaneously sequenced in parallel. The millions of sequenced reads are then aligned against the reference genome and the sequences compared (Figure 4). The massive production of parallel sequences generates several reads for each given position of the genome. The number of reads per stretch of DNA is called coverage. A high coverage improves the accuracy of the interpretation of the sequences since it may filter out errors and noise and also facilitates the detection of mutations in tumor samples with a certain contamination of normal cells. One of the major challenges is the development of reliable bioinformatic algorithms to interpret the sequences. This analysis may detect changes in single nucleotides, the presence of small insertions or deletions (indels), or larger structural alterations. A number of reads above or below the mean coverage per region of DNA will inform about the presence of gains, amplifications, and hemizygous or homozygous deletions. In addition, the mapping of a number of reads in two distant regions of the chromosome or even in different chromosomes will indicate the presence of translocations (Figure 4).3
Next generation sequencing. Next generation sequencing technologies may detect point mutations, small insertions or deletions and large structural variants such as amplifications, homozygous, and heterozygous deletions and translocations. In the RNA sequencing, the number of short reads obtained for each transcript is used to quantify the levels of expression levels of the RNA (modified from ref.3 used with permission).
These sequencing studies may be applied to the WG, the whole transcriptome or may be targeted to specific regions of the genome including all coding exons (exome) (Figure 1). Comparison of the sequences in the tumor with the constitutional DNA of the same individual allows the detection of somatic mutations and filters out thousands of individual polymorphisms. The WG sequences of several solid tumors and hematological neoplasms already have been reported. These studies provide a comprehensive view of all types of somatic mutations but also, given the massive amount of information, provide evidence of possible mechanisms involved in the mutational process.86 The sequence of the exome or other specific targeted regions of the genome are alternative methods that minimize the cost and the speed and still provide relevant information in larger series of patients. The methodologies are based on a selective capture of the genomic fragments of interest using tagged complementary oligonucleotides. The sequencing of the transcriptome or RNA sequencing (RNAseq) starts with cDNA derived from mRNA, total RNA, or other RNAs such as microRNAs. This approach allows the quantification of the transcripts similarly to microarray GEP but without the need of a platform with previously known reference genes. In addition, it allows the discovery of potential new fusion transcripts or transcripts with alternative splice forms. Given the power of these methodologies it is possible to foresee that technologies more adjusted to the scale of clinical problems and the development of friendlier bioinformatic tools may find their way into the clinical practice and may substitute for microarray platforms in the near future.
Landscape of Somatic Mutations in Lymphoid Neoplasms
Initial studies in lymphoid neoplasms have started to display a complex panorama of somatic mutations in these tumors. The sequences of the WG, exome, and transcriptome of a large number of lymphoid neoplasms have been already reported including CLL,86, 87, 88, 89 HCL90 FL,91 DLBCL,91, 92, 93 and plasma cell myeloma (PCM).94 These studies have been the starting point for additional functional and clinical investigations that have confirmed the oncogenic potential and clinical impact of some of the findings. The number of somatic mutations in the genome is variable from around 0.8 mutations per MB in CLL to 2.9 in PCM. The number of mutations in coding regions also varies from 5 to 20 in CLL to ∼35 in PCM, indicating the different mutagenic potential in these tumors.86, 94
Although the number of cases examined in most of these tumors is still relatively low, some common patterns are emerging. The profile of mutations in most of these tumor types is characterized by the presence of few mutated genes in large number of cases and a higher number of mutated genes at a very low frequency. Interestingly, HCL and Waldenstrom’s macroglobulinemia (WM) show an opposite scenario with a single-mutated gene in almost all cases, the BRAF V600E mutation in HCL90 and the MYD88 L256P mutation in the majority of WM.93 Although many mutated genes occur at low frequency, they tend to cluster in common pathogenetic pathways. The main pathways involved in each type of tumor also seem different, suggesting that the transforming mechanisms may differ according to tumor types and cell of origin.
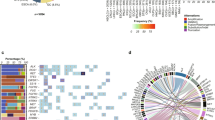
One of the striking surprises of the sequencing studies in CLL has been the large genetic heterogeneity of the disease with a relative large number of genes mutated at low frequency (Figure 5). Only a few mutations are recurrent in 10–15% of the cases. Although some mutations are distributed equally among the IGHV-mutated and -unmutated CLL, other genes appear preferentially mutated in one of the two subtypes. The mutated genes tend to cluster in different pathways that include NOTCH1 signaling, RNA splicing, and processing machinery, inflammatory response, DNA damage and cell-cycle control and WNT pathway among others.87, 88 NOTCH1 mutations, found in ∼10% of CLL, are associated with an adverse prognosis and higher risk of transformation to DLBCL.86, 89 Interestingly, a study of the MCL transcriptome has also found NOTCH1 mutations in 12% of the cases.95 As in CLL, they were associated with a poor prognosis. Mutations in SF3B1, an element of the spliceosome complex, found in ∼10% of CLL, are also associated with a worse outcome. Interestingly, other genes of the RNA splicing and processing machinery are also mutated indicating that this pathway may have a relevant role in the pathogenesis of the disease.87, 88 No mutations in this gene have been detected in other lymphomas but genes of this pathway are frequently mutated in myeloproliferative neoplasms.87, 88
Distribution of different somatic mutations in CLL with mutated and unmutated IGHV.87
The spectrum of mutations in DLBCL is similar to CLL with few genes frequently mutated and a long list of genes mutated at low frequency. However, the targeted pathways are different and some of the genes are altered in a higher proportion of cases than that seen in CLL. For example, the histone methyltransferase MLL2 is mutated in 32% of DLBCL and 89% of FL. The following are the more frequently mutated pathways: chromatin remodeling (EZH2, MLL2, CREBBP, and EP300), immune recognition by T cells (B2M) and post-germinal center differentiation program. Similarly also to CLL, some mutated genes occur in one of the molecular subtypes of DLBCL, ABC, or GCB, whereas others are equally distributed in both categories. Some genes of the BCR signaling and NFκB pathway (CD79b, MYD88, A20) are more commonly mutated in ABC, whereas BCL2 or the methyltransferase EZH2 mutations are mainly found in the GCB subtype.91, 92, 96
PCM has a high mutational load compared with CLL. Interestingly, the mutated genes belong to the protein translation machinery including genes of the unfolded protein responses, a mechanism closely related to the normal secretory function of plasma cells. NFκB and histone-modifying enzymes are also a target of recurrent mutations.94 The landscape of somatic mutations in T-cell neoplasias are starting to emerge. Based on NGS studies, STAT3 mutations have been found in 40% of large granular lymphocytic leukemia. These mutations seem to activate the downstream STAT3 pathway and the patients present more often with neutropenia and rheumatoid arthritis.97
How all this expanding information may be translated into clinical practice is difficult to foresee at the present time. It is still too early to start making predictions but it seems, from the functional and clinical studies already performed for some of the genes, that this new information will have an important impact. On the other hand, many of the mutated genes found were previously unknown in cancer and, therefore, their role in oncogenesis is uncertain. The frequency of some of the apparently relevant mutations seems too low to design specific studies but curiously, for some of them, the mutated gene is usually found across different entities although at different frequencies. For instance, BRAF is mutated in all HCL but also in 2% of the CLL and 4% of PCM.87, 90, 94 Some of the mutated genes, such as NOTCH1 or BRAF, already have experimental drugs available for other tumors in which the mutations were previously found. It will be necessary to study whether these drugs, such as BRAF inhibitors, may be useful in these different entities.
In conclusion, the last decade may be considered the first postgenomic era of hematopathology that, thanks to new technologies, has generated new knowledge with profound impact in our understanding of lymphoid neoplasms. Some of this information has been translated into the clinic but other aspects are still difficult and will require further studies probably with alternative methodologies and standardized procedures. The new generation of sequencing technologies is opening new perspectives with a comprehensive view of the mutational landscape of tumors that may have a clinical impact as predictive biomarkers for new therapies and in certain cases molecular diagnostic markers.
References
Nielander I, Bug S, Richter J, et al. Combining array-based approaches for the identification of candidate tumor suppressor loci in mature lymphoid neoplasms. APMIS 2007;115:1107–1134.
Maciejewski JP, Tiu RV, O’Keefe C . Application of array-based whole genome scanning technologies as a cytogenetic tool in haematological malignancies. Br J Haematol 2009;146:479–488.
Meyerson M, Gabriel S, Getz G . Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 2010;11:685–696.
Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature 2006;444:444–454.
Krijgsman O, Israeli D, Haan JC, et al. CGH arrays compared for DNA isolated from formalin-fixed, paraffin-embedded material. Genes Chromosomes Cancer 2012;51:344–352.
Heinrichs S, Li C, Look AT . SNP array analysis in hematologic malignancies: avoiding false discoveries. Blood 2010;115:4157–4161.
Hosking FJ, Dobbins SE, Houlston RS . Genome-wide association studies for detecting cancer susceptibility. Br Med Bull 2011;97:27–46.
Makishima H, Maciejewski JP . Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res 2011;17:3913–3923.
Hartmann EM, Campo E, Wright G, et al. Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling. Blood 2010;116:953–961.
Swerdlow S, Campo E, Harris N et al (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC: Lyon,, 2008.
Bea S, Campo E . Secondary genomic alterations in non-Hodgkin’s lymphomas: tumor-specific profiles with impact on clinical behavior. Haematologica 2008;93:641–645.
Ferreira BI, Garcia JF, Suela J, et al. Comparative genome profiling across subtypes of low-grade B-cell lymphoma identifies type-specific and common aberrations that target genes with a role in B-cell neoplasia. Haematologica 2008;93:670–679.
Seto M, Honma K, Nakagawa M . Diversity of genome profiles in malignant lymphoma. Cancer Sci 2010;101:573–578.
Bea S, Salaverria I, Armengol L, et al. Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood 2009;113:3059–3069.
Royo C, Salaverria I, Hartmann EM, et al. The complex landscape of genetic alterations in mantle cell lymphoma. Semin Cancer Biol 2011;21:322–334.
Gunnarsson R, Mansouri L, Isaksson A, et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica 2011;96:1161–1169.
Ouillette P, Collins R, Shakhan S, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood 2011;118:3051–3061.
Rinaldi A, Mian M, Chigrinova E, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood 2011;117:1595–1604.
Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA 2008;105:13520–13525.
Salido M, Baro C, Oscier D, et al. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: a multicenter study of the Splenic B-Cell Lymphoma Group. Blood 2010;116:1479–1488.
Fujiwara SI, Yamashita Y, Nakamura N, et al. High-resolution analysis of chromosome copy number alterations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified, with single nucleotide polymorphism-typing microarrays. Leukemia 2008;22:1891–1898.
Salaverria I, Bea S, Lopez-Guillermo A, et al. Genomic profiling reveals different genetic aberrations in systemic ALK-positive and ALK-negative anaplastic large cell lymphomas. Br J Haematol 2008;140:516–526.
Zettl A, Ott G, Makulik A, et al. Chromosomal gains at 9q characterize enteropathy-type T-cell lymphoma. Am J Pathol 2002;161:1635–1645.
Ko YH, Karnan S, Kim KM, et al. Enteropathy-associated T-cell lymphoma—a clinicopathologic and array comparative genomic hybridization study. Hum Pathol 2010;41:1231–1237.
Bea S, Lopez-Guillermo A, Ribas M, et al. Genetic imbalances in progressed B-cell chronic lymphocytic leukemia and transformed large-cell lymphoma (Richter’s syndrome). Am J Pathol 2002;161:957–968.
Tagawa H, Karube K, Tsuzuki S, et al. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci 2007;98:1482–1490.
Navarro A, Bea S, Fernandez V, et al. MicroRNA expression, chromosomal alterations, and immunoglobulin variable heavy chain hypermutations in Mantle cell lymphomas. Cancer Res 2009;69:7071–7078.
Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood 2009;114:2467–2475.
Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009;459:717–721.
Karube K, Nakagawa M, Tsuzuki S, et al. Identification of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK-cell neoplasms by genomic and functional analyses. Blood 2011;118:3195–3204.
Rui L, Emre NC, Kruhlak MJ, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell 2010;18:590–605.
Melzner I, Bucur AJ, Bruderlein S, et al. Biallelic mutation of SOCS-1 impairs JAK2 degradation and sustains phospho-JAK2 action in the MedB-1 mediastinal lymphoma line. Blood 2005;105:2535–2542.
Bignell GR, Greenman CD, Davies H, et al. Signatures of mutation and selection in the cancer genome. Nature 2010;463:893–898.
Iqbal J, Liu Z, Deffenbacher K, et al. Gene expression profiling in lymphoma diagnosis and management. Best Pract Res Clin Haematol 2009;22:191–210.
Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 2006;24:1151–1161.
Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–511.
Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 2003;3:185–197.
Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–1947.
Rosenwald A, Wright G, Leroy K, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 2003;198:851–862.
Savage KJ, Monti S, Kutok JL, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood 2003;102:3871–3879.
Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359:2313–2323.
Said JW . Aggressive B-cell lymphomas—how many categories do we need? Mod Pathol 2012, (this issue).
Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005;105:1851–1861.
Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med 2001;194:1639–1647.
Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med 2001;194:1625–1638.
Basso K, Liso A, Tiacci E, et al. Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhesion receptors. J Exp Med 2004;199:59–68.
Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med 2004;351:2159–2169.
Martinez-Climent JA, Alizadeh AA, Segraves R, et al. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood 2003;101:3109–3117.
Glas AM, Kersten MJ, Delahaye LJ, et al. Gene expression profiling in follicular lymphoma to assess clinical aggressiveness and to guide the choice of treatment. Blood 2005;105:301–307.
Fu K, Weisenburger DD, Greiner TC, et al. Cyclin D1-negative mantle cell lymphoma: a clinicopathologic study based on gene expression profiling. Blood 2005;106:4315–4321.
Fernandez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res 2010;70:1408–1418.
Navarro A, Clot G, Royo C, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biological and clinical features. Cancer Res 2012, (in press).
Arribas JA, Campos-Martin Y, Gomez-Abad C, et al. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood 2012;119:e9–e21.
Novak U, Basso K, Pasqualucci L, et al. Genomic analysis of non-splenic marginal zone lymphomas (MZL) indicates similarities between nodal and extranodal MZL and supports their derivation from memory B-cells. Br J Haematol 2011;155:362–365.
Iqbal J, Weisenburger DD, Greiner TC, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood 2010;115:1026–1036.
Piccaluga PP, Agostinelli C, Califano A, et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res 2007;67:10703–10710.
de Leval L, Rickman DS, Thielen C, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 2007;109:4952–4963.
Lamant L, de RA, Duplantier MM, et al. Gene-expression profiling of systemic anaplastic large-cell lymphoma reveals differences based on ALK status and two distinct morphologic ALK+ subtypes. Blood 2007;109:2156–2164.
Iqbal J, Weisenburger DD, Chowdhury A, et al. Natural killer cell lymphoma shares strikingly similar molecular features with a group of non-hepatosplenic gammadelta T-cell lymphoma and is highly sensitive to a novel aurora kinase A inhibitor in vitro. Leukemia 2011;25:348–358.
Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med 2006;354:2431–2442.
Harris NL . The bridge from the large B-cell lymphomas to Hodgkin lymphomas: differential diagnosis and biological implications. Mod Pathol 2012, (this issue).
Traverse-Glehen A, Pittaluga S, Gaulard P, et al. Mediastinal gray zone lymphoma: the missing link between classic Hodgkin’s lymphoma and mediastinal large B-cell lymphoma. Am J Surg Pathol 2005;29:1411–1421.
Eberle FC, Salaverria I, Steidl C, et al. Gray zone lymphoma: chromosomal aberrations with immunophenotypic and clinical correlations. Mod Pathol 2011;24:1586–1597.
Quintanilla-Martinez L, Fend F . Mediastinal gray zone lymphoma. Haematologica 2011;96:496–499.
Eberle FC, Rodriguez-Canales J, Wei L, et al. Methylation profiling of mediastinal gray zone lymphoma reveals a distinctive signature with elements shared by classical Hodgkin’s lymphoma and primary mediastinal large B-cell lymphoma. Haematologica 2011;96:558–566.
Oschlies I, Burkhardt B, Salaverria I, et al. Clinical, pathological and genetic features of primary mediastinal large B-cell lymphomas and mediastinal gray zone lymphomas in children. Haematologica 2011;96:262–268.
Carreras J, Villamor N, Colomo L, et al. Immunohistochemical analysis of ZAP-70 expression in B-cell lymphoid neoplasms. J Pathol 2005;205:507–513.
Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med 2003;348:1764–1775.
Ek S, Dictor M, Jerkeman M, et al. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood 2008;111:800–805.
Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica 2009;94:1555–1562.
Ondrejka SL, Lai R, Kumar N, et al. Indolent mantle cell leukemia: clinicopathologic variant characterized by isolated lymphocytosis, interstitial bone marrow involvement, kappa light chain restriction, and good prognosis. Haematologica 2011;96:1121–1127.
Cuadros M, Dave SS, Jaffe ES, et al. Identification of a proliferation signature related to survival in nodal peripheral T-cell lymphomas. J Clin Oncol 2007;25:3321–3329.
Ngo VN, Davis RE, Lamy L, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 2006;441:106–110.
Lam LT, Wright G, Davis RE, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008;111:3701–3713.
Scuto A, Kujawski M, Kowolik C, et al. STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res 2011;71:3182–3188.
Dunleavy K, Pittaluga S, Czuczman MS, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood 2009;113:6069–6076.
Reeder CB, Ansell SM . Novel therapeutic agents for B-cell lymphoma: developing rational combinations. Blood 2011;117:1453–1462.
Williams PM, Li R, Johnson NA, et al. A novel method of amplification of FFPET-derived RNA enables accurate disease classification with microarrays. J Mol Diagn 2010;12:680–686.
Hartmann E, Fernandez V, Moreno V, et al. Five-gene model to predict survival in mantle-cell lymphoma using frozen or formalin-fixed, paraffin-embedded tissue. J Clin Oncol 2008;26:4966–4972.
Rimsza LM, Wright G, Schwartz M, et al. Accurate classification of diffuse large B-cell lymphoma into germinal center and activated B-cell subtypes using a nuclease protection assay on formalin-fixed, paraffin-embedded tissues. Clin Cancer Res 2011;17:3727–3732.
Alizadeh AA, Gentles AJ, Alencar AJ, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood 2011;118:1350–1358.
Hoster E, Dreyling M, Klapper W, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood 2008;111:558–565.
De Jong D, Rosenwald A, Chhanabhai M, et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications—a study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol 2007;25:805–812.
De Jong D, Koster A, Hagenbeek A, et al. Impact of the tumor microenvironment on prognosis in follicular lymphoma is dependent on specific treatment protocols. Haematologica 2009;94:70–77.
De Jong D, Wahlin B, Sander B, et al. The reliability of immunohistochemical analysis of tumor microenvironment in follicular lymphoma; a validation study from the lunenburg lymphoma biomarker consortium (LLBC). Ann Oncol 2011;22:108.
Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011;475:101–105.
Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2011;44:47–52.
Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011;365:2497–2506.
Fabbri G, Rasi S, Rossi D, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med 2011;208:1389–1401.
Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med 2011;364:2305–2315.
Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011;476:298–303.
Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet 2011;43:830–837.
Treon SP, Xu L, Zhou Y, et al. Whole Genome Sequencing Reveals a Widely Expressed Mutation (MYD88 L265P) with Oncogenic Activity in Waldenstrom’s Macroglobulinemia. 53rd Annual Meeting and Exposition of the American-Society-of-Hematology (ASH) [abstract]. Blood 2011;118:140.
Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011;471:467–472.
Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 2012;119:1963–1971.
Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA 2012;109:3879–3884.
Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med 2012;366:1905–1913.
Acknowledgements
I thank Dr SH Swerdlow for inviting me to share the responsibility to plan and organize this Long Course. It has been a unique and creative experience that, thanks also to the excellent contributions of all the speakers and participants, we hope will enrich our speciality and prepare all of us for the challenges of the future. I would like to thank my colleagues Sílvia Beà and Pedro Jares for their comments and help in the preparation of the manuscript and all the members of the Spanish ICGC-CLL Genome Project, particularly to Dr Carlos Lopez-Otin, for their commitment and instrumental collaboration. Part of the own work presented here has been funded by the Spanish Ministry of Science and Innovation and Instituto de Salud Carlos III (ICGC-CLL genome project; SAF 2008-03630).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Campo, E. Whole genome profiling and other high throughput technologies in lymphoid neoplasms—current contributions and future hopes. Mod Pathol 26 (Suppl 1), S97–S110 (2013). https://doi.org/10.1038/modpathol.2012.179
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2012.179