Abstract
We have previously established immortalized cells (HCF) from cementifying fibroma of the jaw bone. Here, we found that the receptor for hyaluronan (HA)-mediated motility (RHAMM) and epiregulin, a ligand for the epidermal growth factor receptor (EGFR), were highly expressed in HCF cells in comparison with osteoblasts by conducting a microarray analysis. The cell growth of HCF cells was significantly decreased by the knockdown of RHAMM using small interfering RNA (siRNA). RHAMM was associated with extracellular signal-regulated kinase (ERK) and essential for ERK phosphorylation. HCF cells had characteristic growth mechanisms in which epiregulin functions in an extracellular autocrine loop. Interestingly, exogenous HA induced the phosphorylation of EGFR, which was mainly dependent on CD44. The results raise the novel idea that the EGFR may activate Raf–MEK–ERK signaling in response to the binding of HA to CD44. Moreover, RHAMM was able to associate with TPX2 in the nucleus and was required for HA-induced activation of the Aurora A kinase. The results suggest that RHAMM has a predominant role in the cell cycle in HCF. Here, we report the new machinery by which RHAMM/ERK interaction induces the proliferative activity of cementifying fibroma cells via a specific signaling pathway through the CD44–EGFR axis.
Similar content being viewed by others
Main
Benign fibro-osseous diseases in the oral and maxillofacial regions can be divided into three categories: fibrous dysplasia, osseous dysplasia, and cementifying (ossifying) fibroma.1, 2 Cementifying fibroma is composed of fibrous tissue containing varying amounts of mineralized tissues resembling cementum or bone. Recently, we established immortalized cell lines (HCF) from human cementifying fibroma of the jaw.3 We performed the double transfection of simian virus-40 (SV40) T-antigen and h-TERT to obtain the immortalized cells.3 The mechanisms of cellular differentiation and proliferation in cementifying fibroma cells have not been elucidated. To identify the characteristic gene expression in HCF cells, a microarray analysis was performed. We found that several genes were highly expressed compared with normal human osteoblasts. Among them, we found that the receptor for hyaluronan (HA)-mediated motility (RHAMM) was highly expressed in HCF cells.
RHAMM has been identified as a cell-surface receptor for HA.4, 5, 6, 7 Human RHAMM cDNA was cloned and contained 725 amino acids that encoded an 85-kDa protein.4 Other HA-binding proteins have been identified, and CD44 has been characterized as a molecularly distinct cell-surface receptor for HA.8, 9, 10 HA is a glycosaminoglycan composed of repeating disaccharide units of D-glucuronic acid and N-acetyl-D-glucosamine and an important component of the extracellular matrix.11 HA has been reported to stimulate proliferation in various types of cells.12, 13, 14, 15 The binding of HA to RHAMM generates transmembrane signals that trigger signaling cascades leading to cytoskeletal rearrangements.9 RHAMM seems to mediate HA-stimulated proliferation and migration not only in vascular smooth muscle cells but also in tumor cells.16, 17, 18
The cell growth mediated by RHAMM is considered to occur via signaling events leading to the phosphorylation of several intracellular proteins.19 Moreover, RHAMM is thought to control extracellular signal-regulated kinase (ERK), which has a significant role in cell motility and cellular proliferation.20, 21 However, the mechanisms by which RHAMM controls signal transduction in mammalian cells have not been fully elucidated. Recently, we identified that RHAMM was overexpressed in oral squamous cell carcinomas.22 The overexpression of RHAMM has also been observed in other types of cancers.23, 24, 25 In oral squamous cell carcinomas, RHAMM expression was correlated with the expression of TPX2 (targeting protein for X. laevis kinesin-like protein 2), encoding a protein which is required for the formation of microtubules at kinetochores in mammalian cells.22 RHAMM interacted with TPX2 in multiple myeloma cells.26 TPX2 initiates spindle assembly by activating the Aurora A kinase in a microtubule-dependent manner.27 Collectively, previous research indicates that RHAMM has an initial role in regulating the integrity of mitotic-spindle assemblies in concert with TPX2 and induces cellular proliferation under malignant conditions. However, the initial biological behavior of RHAMM has not been fully investigated in benign tumors including cementifying fibroma.
In an effort to clarify the possible and specific growth mechanism in cementifying fibroma, we have further focused on the expression of epiregulin, a novel ligand of the epidermal growth factor receptor (EGFR).28 The binding of this ligand to the EGFR promotes activation of the tyrosine kinase function. Subsequently, autophosphorylation of the receptor activates signaling pathways that regulate proliferation and cell survival. We identified the participation of extracellular epiregulin autocrine loops involving the EGFR in promoting Raf–MEK–RHAMM/ERK signaling in HCF cells. In this paper, we report the identification of a specific growth mechanism in HCF cells and highlight the novel signaling through RHAMM/ERK interaction.
MATERIALS AND METHODS
Cell Culture
Immortalized HCF cell lines were established by co-transfection with SV40 T-antigen and h-TERT.3 We used one clone of the HCF cell lines in this study. The HCF cells were maintained in α-modified Eagle medium (α-MEM; Sigma) supplemented with penicillin/streptomycin and 10% heat-inactivated fetal bovine serum (FBS; Invitrogen) under 5% CO2 in air at 37 °C. They were then cultured with the same medium and used for the following analyses. TK-1 and SK-1 cells were human osteoblasts obtained from normal human mandibular bone and cultured with α-MEM. HF-1 cells were fibroblastic cells previously established from normal human gingiva in our laboratory. GT-1 cells were human gingival fibroblasts immortalized by transfection with h-TERT.29 These cells were maintained in DMEM supplemented with penicillin/streptomycin and 10% heat-inactivated FBS under 5% CO2 in air at 37 °C. The culture medium was changed every 3 days. To determine the effects of HA or epiregulin, HCF cells were plated into six-well plates at a density of 5 × 104 cells/ml and allowed to attach for 24 h. To determine the effect of HA, HCF cells were incubated with a synthesized 160-kDa HA (Hyalose). To examine the effect of exogenous epiregulin, cells were exposed to medium containing 100 ng/ml of a recombinant epiregulin (R&D Systems). To ascertain the effect of HA or epiregulin, cells were exposed to the medium with 1% FBS. Cells were trypsinized, and the number of cells was counted using a cell counter (Coulter Z1; Coulter). Anti-EGFR antibody (Thermo Fisher Scientific) was used for blocking EGF/TGF-induced activation. EGFR inhibitor (Calbiochem) for blocking the tyrosine kinase activity of EGFR was used at a final concentration of 10 μM.
Staining for Mineralization
Mineralization of the cells was also examined by staining with 1% Alizalin red S (Wako Pure Chemicals Industries). After reaching confluence, cells were cultured for 3 more weeks in the presence of 15 mM β-glycerophosphate and 100 μg/ml ascorbic acid, and the activity to form a mineralized matrix was examined. Alizalin red S-stained samples were examined under a dissecting microscope to identify calcium phosphate deposits. The stain was extracted with 10% cetylpyridium chloride in 10 mM sodium phosphate. The absorbance at 575 nm was read using a Microplate reader model 680 (Bio-Rad).
Quantitative RT–PCR Analysis
The quantification of mRNA levels was carried out using a real-time fluorescence detection method. The fluorescence was detected by the laser detector of a Line Gene Fluorescent Quantitative Detection System (Bio Flux), and detection was carried out by measuring the binding of a fluorescence dye, SYBR Green I, to double-stranded DNA. The reaction mixture contained 1.0 μg of cDNA, 10 μl of SYBR Green PCR Master Mix (Toyobo), and 10 pmol of each pair of oligonucleotide primers. The primer sequences were: RHAMM; 5′-TCATCATTACAGGAAGAAGCG-3′ (sense), 5′-CTGTTGCTTGAGTTGGTTCTG-3′ (antisense), and G3PDH; 5′-ACCACAGTCCATGCCATCAC-3′ (sense), 5′-TCCACCACCCTGTGGCTGTA-3′ (antisense). The primer sequences for alkaline phosphatase (ALP) were described previously.30 The PCR program was as follows: initial melting at 95 °C for 30 s followed by 40 cycles at 95 °C for 15 s, 55 °C for 10 s, and 72 °C for 15 s. The quantification of mRNA expression relative to an internal control, G3PDH, was performed by the ΔCt method.31
Microarray Analysis
The microarray analysis was performed with a Human 35K Oligo DNA Micro Array (Filgen) containing 34 580 gene spots of 70-mer oligonucleotides. Hybridization was performed for 16 h at 42 °C in 80 μl of a solution containing 5 × SCC, 0.1% sodium dodecyl sulfate (SDS), 10% formamide, and heat-denatured labeled cDNA. The fluorescence images of Cy3 and Cy5 were obtained with a GenePix 4000B scanner (Axon Instruments). The equation was revised based on an average of the intensity on one filter (global normalization) using an Array-ProAnalyzer (Media Cybernetics), and normalization of the experiments was performed with the Lowess method.32
Western Blotting
Protein concentrations were measured using the protein assay reagent (Bio-Rad). Protein samples (10 μg) were solubilized in sample buffer by boiling, subjected to SDS–polyacrylamide gel electrophoresis (PAGE) followed by electorotransfer onto a nitrocellulose filter (Schleicher & Schuell). We detected the band on western blotting using enhanced chemiluminescence (ECL) western blotting reagent (GE Healthcare). Images were captured with a cooled CCD camera system (LAS-4000) from Fujifilm. An anti-human epiregulin mouse monoclonal anti-body (Chemicon), anti-human RHAMM goat polyclonal anti-body (Santa Cruz Biotechnology), anti-human ERK1/2 rabbit polyclonal antibody (Cell Signaling Technology), anti-human MEK1/2 rabbit polyclonal antibody (Cell Signaling Technology), anti-human Raf-1 rabbit polyclonal antibody (Cell Signaling Technology), anti-human Aurora A/B rabbit polyclonal antibody (Cell Signaling Technology), anti-human phosphorylated-Aurora A/B (Thr 288/Thr 232) rabbit polyclonal antibody (Cell Signaling Technology), anti-human TPX2 monoclonal antibody (Santa Cruz Biotechnology), anti-human CD44 monoclonal antibody (Santa Cruz Biotechnology), and anti-human phosphorylated EGFR (Tyr 1068) rabbit polyclonal antibody (Cell Signaling Technology) were used.
Immunoprecipitation Assay
HCF cells were treated with or without 100 μg/ml HA for 1 h. Antibodies were allowed to bind to protein A-Sepharose (Sigma-Aldrich) and then incubated with equal amounts of protein (0.5 mg of total protein in 400 μl) for 12 h at 4 °C. Beads were washed three times with PBS. Each pellet was boiled in 20 μl of SDS–polyacrylamide gel electrophoresis sample buffer at 95 °C for 3 min, and the entire volume was loaded onto a gel for western blotting.
Small Interfering RNA
The Stealth™ small interfering RNA (siRNA) for RHAMM (BC108904) and the negative control siRNA were purchased from Invitrogen. HCF cells were transiently transfected with the indicated combinations of the siRNAs using Lipofectamine™ 2000 transfection reagent (Invitrogen), according to the manufacturer's recommendations. Two independent RNA oligos were used for the knockdown of RHAMM in order to rule out off-the-target effects. We confirmed the same results of each experiment using two independent siRNAs.
Hyarulonan-Binding Assay
Hyarulonan-binding assay was performed to determine whether RHAMM can bind HA as described previously.33 Cell lysate was centrifuged, and aliquots (100 μl) were mixed with 50 μl of GAG (40 μg of HA, chondroitin sulfate, dermatan sulfate, heparin sulfate, heparin, and dermatan sulfate). After incubation at room temperature for 1 h, 350 μl of 1.4% cetylpyridinium chloride (CPC) was added to the mixture of lysate and GAG. After incubation for 1 h, the mixture was centrifuged at maximum speed for 15 min. The pellets were washed with 1% CPC containing 20 mM NaCl and dissolved in SDS–polyacrylamide gel electrophoresis sample buffer at 95 °C for 3 min. The supernatant was analyzed by western blotting. GAG was used as a negative control for the western blotting.
Immunofluorescence Microscopy
HCF cells were seeded onto glass Lab-Tek II Chamber Slides (Thermo Fisher Scientific) at a density of 5.0 × 104 cells/well and incubated for a day. The cells were exposed to medium containing HA (100 μg/ml) for 1 h. The growth medium was then removed, and cell monolayers were washed three times with a 10% PBS solution and fixed with 3.5% paraformaldehyde for 10 min at room temperature. Cells were washed three times with PBS and permeabilized by Triton X-100 (0.2%, 10 min at room temperature). Nonspecific binding sites were blocked by treatment at room temperature for 30 min with PBS containing 1% BSA. The cells were washed three times with PBS and incubated with anti-RHAMM mouse monoclonal antibody (MONOSAN) (1:100)/anti-TPX2 rabbit polyclonal antibody (Lifespan Biosciences) (1:100) in PBS with 1% BSA, for 60 min at room temperature. RHAMM staining was revealed by incubation with an Alexa-Fluor dye-labeled goat anti-mouse antibody (1:40; Invitrogen) for 60 min at room temperature. TPX2 staining was revealed by incubation with an Alexa-Fluor dye-labeled goat anti-rabbit antibody (1:40; Invitrogen) for 60 min at room temperature. After three rinses in PBS, the slides were mounted in Vectashield (Vecto Laboratories) and examined using a Leica TCS STED (Leica Microsystems).
Cell Cycle Analyses
The distribution of cells at different stages in the cell cycle was estimated by flow cytometric analysis. Briefly, 5 × 105 cells were incubated at 37 °C in triplicate 10-cm plastic dishes. To synchronize them in G0, the HCF cells were starved by serum deprivation for 48 h. After restimulation with medium containing 10% FBS, cells passed through the cell cycle synchronously and were incubated for 24 h/48 h. Cells from the different conditions were trypsinized, washed in PBS, fixed in 70% ethanol, and stored at 4 °C for 2 h. An aliquot (1 ml) of the fixed cell suspension was washed twice in PBS. The fixed cells were treated for 30 min at 4 °C in the dark with 40 μg of propidium iodide and 0.1 mg of RNase A, and then analyzed by flow cytometry. The percentage of cells in each cell cycle phase (G0/G1, S, or G2/M) was calculated by using ModFit LT Software (Becton Dickinson).
Statistical Methods
The statistical analysis was performed using the one-way ANOVA and Student's t-test. P-values <0.05 were regarded as statistically significant.
RESULTS
RHAMM and Epiregulin Are Highly Expressed in HCF Cells
The microarray analysis identified 26 genes whose expression was >6.0-fold higher in HCF cells than SK-1 cells (Table 1). As the SK-1 cells were osteoblasts derived from normal human jaw bone, we used them as a normal control for HCF cells. RHAMM mRNA was expressed at 6.06-fold higher levels in HCF than SK-1 cells. Epiregulin mRNA was also highly expressed at 8.06-fold higher levels in HCF cells (Table 1). Epiregulin is a ligand for the EGFR.28 Expression of RHAMM and epiregulin protein in HCF cells was confirmed by western blotting (Figure 1). EGFR protein expression was detected in HCF cells.
RHAMM Associates with ERK and Is Essential for ERK Phosphorylation
To determine whether RHAMM is necessary for the growth of HCF cells, we performed the siRNA-mediated knockdown of RHAMM. After elimination of the gene by siRNA, the protein expression was clearly suppressed in HCF cells (Figure 2a). The cell growth was significantly decreased by the knockdown of RHAMM compared with the control (*P<0.01) (Figure 2b). In an effort to identify the mechanisms by which the RHAMM-mediated cell growth occurs, we focused on the activation of Raf–MEK–ERK signaling. Western blotting revealed that levels of phosphorylated Raf-1 and MEK1/2 in RHAMM-knockdown cells were not attenuated in comparison with the control cells (Figure 2c). On the other hand, RHAMM knockdown inhibited phosphorylation of ERK1/2 (Figure 2c). The results suggest RHAMM may have an important role in the phosphorylation of ERK. To determine whether RHAMM can associate with ERK1/2, an immunoprecipitation assay was performed. Cell lysate pretreated with HA was immunoprecipitated with the anti-RHAMM antibody or anti-ERK1/2 antibody and immunoblotted. As shown in Figure 2d, RHAMM was found to be associated with ERK1/2 in the presence of HA. ERK1/2 was also found to be associated with RHAMM in an immunoprecipitation assay for ERK1/2 (Figure 2d). The immunoprecipitation assay for ERK revealed increased phosphorylation of ERK1/2 in the presence of HA. RHAMM is thought to associate with and function as an activator of ERK in other types of cells.20, 21 Our results support the notion that RHAMM can associate with ERK and is essential for activating ERK1/2 in HCF and other cells.
RHAMM can associate with ERK and is essential for ERK phosphorylation. (a) Small interfering RNA (siRNA) of RHAMM in HCF cells. RHAMM expression was decreased in the RHAMM-knockdown HCF cells by western blotting. (b) Growth inhibition by the RHAMM knockdown in HCF cells. The cell growth was significantly inhibited by the RHAMM-knockdown (*P<0.01). The results are expressed as the mean±s.d. for three independent cultures. (c) Levels of phosphorylated Raf-1, MEK1/2, and ERK1/2 in the RHAMM-knockdown cells. Phosphorylation of ERK1/2 was decreased by the RHAMM knockdown. (d) Immunoprecipitation assay for the analysis of RHAMM and ERK complexes in HCF cells. Goat IgG was used as a negative control for the RHAMM goat polyclonal antibody. Rabbit IgG was used as a negative control for the ERK1/2 rabbit polyclonal antibody. RHAMM was associated with ERK1/2 in the presence or absence of HA in immunoprecipitation assays for RHAMM. Immunoprecipitation assays showed that ERK was associated with RHAMM. Phosphorylation of ERK1/2 was increased in the presence of HA.
RHAMM Is Required for Epiregulin-Induced Activation of ERK
To clarify the effect on growth in the presence of exogenous epiregulin, the cells were exposed to medium with 1% FBS containing a recombinant epiregulin for 7 days. Maximal growth of the HCF cells was observed in the presence of 100 ng/ml of epiregulin (data not shown). The numbers of HCF cells were significantly increased in the epiregulin (100 ng/ml)-added medium compared with the control at 7 days (*P<0.01) (Figure 3a). Furthermore, levels of phosphorylated EGFR, Raf, MEK, and ERK were increased after 60 min in the presence of epiregulin, and inhibited by the addition of the anti-EGFR antibody or EGFR inhibitor (Figure 3b). The phosphorylation of the EGFR, Raf, MEK, and ERK was evaluated by comparing the integrated density of phosphorylated bands on western blots. We performed densitometric scanning using Kodak Digital Science 1D Software (Kodak) and calculated the levels of phosphorylated EGFR, Raf, MEK, and ERK in comparison with the control values. The epiregulin-induced phosphorylation of EGFR, Raf, MEK, and ERK was significantly inhibited by the anti-EGFR antibody and the EGFR inhibitor (*P<0.01) (Figure 4). These results suggested that epiregulin could function as a ligand of the EGFR and induce cell proliferation via the Raf–MEK–ERK pathway. In the RHAMM knockdown cells, the epiregulin-induced phosphorylation of ERK was also decreased (Figure 3b). The epiregulin-triggered phosphorylation of ERK1/2 was significantly blocked by the knockdown of RHAMM (*P<0.01) (Figure 4).
Stimulation of growth by epiregulin in HCF cells. (a) Epiregulin induces cell proliferation in HCF cells. HCF cells were cultured in the presence of epiregulin (100 ng/ml) with 1% FBS for 7 days. The numbers of HCF cells were significantly increased in the presence of epiregulin (100 ng/ml) compared with the control at 7 days (*P<0.01). The anti-EGF receptor antibody blocked the growth of the HCF cells in the presence of epiregulin (*P<0.01). The results are expressed as the mean±s.d. for three independent cultures. (b) Levels of phosphorylated EGFR, Raf-1, MEK1/2, and ERK1/2 in the presence of epiregulin. Cells were exposed to medium containing epiregulin (100 ng/ml) for 1 h. Levels of phosphorylated Raf-1, MEK1/2, and ERK1/2 were decreased in the anti-EGFR antibody and EGFR inhibitor-added cells compared with epiregulin-added cells. Epiregulin-induced phosphorylation of ERK1/2 was inhibited by the RHAMM knockdown.
Quantitative analysis of the phosphorylation of EGFR, Raf-1, MEK1/2, and ERK1/2 in the presence of epiregulin. The levels of phosphorylated EGFR, Raf, MEK, and ERK were evaluated by the density of phosphorylated bands on western blot and calculated in comparison with the control values (phosphorylation level: 1.0). The results are expressed as the mean±s.d. Epiregulin-induced phosphorylation of ERK1/2 was significantly blocked by the RHAMM knockdown (*P<0.01).
RHAMM seems to act as an activator of ERK in signaling downstream of the EGFR. Collectively, these results indicate that epiregulin operates in an extracellular autocrine loop via EGFR, so that the receptor can be activated constitutively in HCF cells.
RHAMM Is Required for HA-Induced Phosphorylation of ERK Through a Mechanism Based on the CD44–EGFR
CD44 is thought to function as a receptor for HA and to have a major role in HA-mediated signaling. We confirmed that not only CD44 but also RHAMM was able to bind to exogenous HA in hyarulonan-binding assays (Supplementary Figure 1a). To ascertain the effect on the growth in the presence of HA, the cells were exposed to the medium with 1% FBS for 7 days. Maximal growth of the HCF cells was observed at 10 μg/ml of HA (data not shown). The growth was significantly increased under HA (10 μg/ml)-added conditions compared with the control after 7 days (*P<0.01) (Figure 5a). HA-induced cell growth was significantly blocked by the knockdown of RHAMM (*P<0.01) as well as in the presence of the anti-CD44 antibody (*P<0.01) (Figure 5a). Interestingly, HA-induced cell growth was also inhibited by the EGFR inhibitor (*P<0.01) (Figure 5a). The results raised the possibility that activated EGFR contributes to the HA-induced growth of HCF cells.
HA-induced EGFR phosphorylation is blocked by inhibition of CD44. (a) HA-induced growth of HCF cells. Cell growth was significantly increased in the presence of 10 μg/ml of HA compared with the control after 7 days (*P<0.01). HA-induced cell growth was significantly inhibited by RHAMM knockdown (*P<0.01). HA-induced cell growth was also significantly inhibited by the anti-CD44 antibody or EGFR inhibitor (*P<0.01). The results are expressed as the mean±s.d. for three independent cultures. (b) The level of EGFR phosphorylation was increased at 30 min after HA treatment. (c) HA-induced phosphorylation of the EGFR was suppressed by the anti-CD44 antibody. Quantitative analysis of the western blotting results revealed that the phosphorylation of the EGFR was significantly decreased in the presence of the anti-CD44 antibody (*P<0.01) or EGFR inhibitor (*P<0.01). The results are expressed as the mean±s.d. (d) HA-induced phosphorylation of Raf/MEK/ERK in HCF cells. Levels of phosphorylated ERK were increased at 15 min after the addition of HA. (e) HA-induced phosphorylation of Raf-1, MEK1/2, and ERK1/2 was reduced by the EGFR inhibitor or anti-CD44 antibody. HA-induced phosphorylation of ERK1/2 was inhibited by RHAMM knockdown.
To clarify whether the EGFR could be activated in response to exogenous HA, we examined the levels of HA-induced EGFR phosphorylation by western blotting. An elevation in the levels of phosphorylated EGFR was observed at 30 min after the HA treatment (Figure 5b). Densitometric analysis of the western blotting results revealed the phosphorylation of EGFR to be significantly increased in the presence of HA (*P<0.01) (Figure 5c). Moreover, it was significantly decreased in the presence of the anti-CD44 antibody (*P<0.01) or EGFR inhibitor (*P<0.01) (Figure 5c). The results suggested that the HA-induced phosphorylation of the EGFR was significantly suppressed by the anti-CD44 antibody. The activation of EGFR might be mainly provoked by CD44's activation. It is a novel finding that CD44-HA binding modulates the augmentation of the activation of EGFR.
The phosphorylation of Raf, MEK, and ERK was examined in the presence of HA (10 μg/ml) by western blotting (Figure 5d). Levels of phosphorylated Raf-1 and MEK1/2 were increased after 30 min (Figure 5d). The phosphorylation of ERK1/2 was increased at 15 min (Figure 5d). HA-induced phosphorylation of Raf-1, MEK1/2, and ERK1/2 was inhibited by the anti-CD44 antibody (Figure 5e). Densitometric analysis of the western blotting results showed that the levels of phosphorylated Raf-1, MEK1/2, and ERK1/2 were significantly reduced in the presence of the anti-CD44 antibody (*P<0.01) (Figure 6). The results raise the possibility that CD44 is a major receptor for HA and can activate the Raf–MEK–ERK signaling pathway via the activation of EGFR. HA-induced ERK phosphorylation was inhibited by the RHAMM knockdown (Figure 5e). The level of phosphorylated ERK1/2 was significantly reduced by the RHAMM knockdown (*P<0.01) (Figure 6). Consistent with previous data, RHAMM is thought to be involved in the activation of ERK in the presence of HA. RHAMM can associate with ERK and is required for HA-induced phosphorylation of ERK. HA-induced phosphorylation of Raf-1, MEK1/2, and ERK1/2 was inhibited by the EGFR inhibitor (Figure 5e). Moreover, the levels of phosphorylated Raf-1, MEK1/2, and ERK1/2 were significantly inhibited by EGFR inhibitor (*P<0.01) (Figure 6). These results raised the novel idea that EGFR promotes the activation of Raf–MEK–ERK signaling in response to the binding of HA to CD44.
Quantitative analysis of the phosphorylation of Raf-1, MEK1/2, and ERK1/2 in the presence of HA. The levels of phosphorylated Raf, MEK, and ERK were evaluated by the density of phosphorylated bands on western blot and calculated in comparison with the control values (phosphorylation level: 1.0). The results are expressed as the mean±s.d. HA-induced phosphorylation of Raf-1, MEK1/2, and ERK1/2 was significantly inhibited by the EGFR inhibitor (*P<0.01).
RHAMM Associates with TPX2 in the Presence of HA
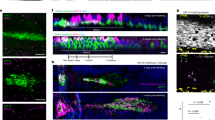
It has been reported that RHAMM interacted with spindle-assembly related protein, TPX2, in myeloma cells.26 Therefore, we performed an immunoprecipitation assay to investigate whether RHAMM can associate with TPX2 in HCF cells. As shown in Figure 7a, the RHAMM protein was found to be mainly associated with TPX2 in the presence of HA by immunoblotting. TPX2 was also found to be associated with RHAMM in response to exogenous HA (Figure 7a). Furthermore, we performed double-immunofluorescence staining of RHAMM and TPX2 to clarify the localization of these proteins. In the absence of HA, RHAMM was mostly observed in the cytoplasm of the cells (Figure 7b). TPX2 was observed in the nucleus in the absence of HA (Figure 7c). The expression of RHAMM and TPX2 is merged in Figure 7d. Interestingly, RHAMM was detected in the centrosome and mitotic spindle of a large number of HCF cells at 60 min after the HA treatment (Figures 7e and h). The treatment of cells with HA resulted in the nuclear accumulation of RHAMM. TPX2 was also detected in the centrosome and mitotic spindle under the same conditions (Figures 7f and i). Co-localization of RHAMM and TPX2 could be found in the nucleus (Figures 7g and j). These results suggest that RHAMM can move to the nucleus immediately and associate with TPX2 at the mitotic spindle and centrosome in response to HA. It is thought that HA can facilitate the distribution of RHAMM in HCF cells.
RHAMM associates with TPX2 and induces the phosphorylation of Aurora A kinase in the presence of HA. (a) Immunoprecipitation assays for RHAMM and TPX2 by immunoblotting. Mouse IgG1 was used as a control isotype IgG for the TPX2 mouse monoclonal antibody. In the presence of HA, RHAMM was mostly associated with TPX2. TPX2 was also associated with RHAMM under the same conditions. (b–j) Immunofluorescence microscopy of RHAMM and TPX2 in HCF cells. (b) RHAMM expression (green) was observed in the cytoplasm of HCF cells. (c) TPX2 expression (red) was observed in nuclei of HCF cells. (d) Merged picture of RHAMM and TPX2. (e) RHAMM expression (green) was observed in the nuclei of HCF cells at 60 min after the addition of HA. (f) TPX2 expression (red) was observed in nuclei of HCF cells. (g) Merged picture of RHAMM and TPX2 shown as an overlay of the two colors (yellow). (h, i) RHAMM and TPX2 expression was observed in the centrosome and mitotic spindle of HCF cells in the presence of HA. (j) Merged picture of RHAMM and TPX2 shown as yellow. (k) Phosphorylation of Aurora A was examined by immunoblotting. The Level of phosphorylated Aurora A was increased at 3 h after the addition of HA, but inhibited by RHAMM knockdown and the anti-CD44 antibody. (l) Quantitative analysis of the western blotting results showed that levels of phosphorylated Aurora A were significantly increased in the presence of HA (*P<0.01) and significantly decreased by RHAMM knockdown/anti-CD44 antibody (*P<0.01). The results are expressed as the mean±s.d. (m) Flow cytometric analysis in HCF cells. The quantitation of the results was performed with ModFit LT Software. The results are presented as the mean±s.d. In HA-added control cells, the percentage of cells in the G2/M phase was 3.8±1.3% at 0 h and 47.9±1.0% at 48 h. In RHAMM-knockdown cells, this percentage was 2.6±0.45% at 0 h and 16.2±0.74% at 48 h. The percentage of cells in the G2/M phase was considerably reduced in RHAMM-knockdown cells compared with control cells at 48 h under HA-added conditions.
RHAMM Is Required for HA-Induced Activation of Aurora A Kinase
We demonstrated that RHAMM could interact with TPX2, which has a significant role in activating spindle-related kinase. To clarify the activities of mitotic spindle-related kinase, we further examined the activation of Aurora A kinase by western blotting. The phosphorylation of Aurora A kinase was increased at 3 h after HA treatment and decreased by the RHAMM knockdown (Figure 7k). Densitometric analysis of the western blotting results showed the levels of phosphorylated Aurora A kinase to be significantly increased in the presence of HA (*P<0.01) and significantly decreased by the knockdown of RHAMM (*P<0.01) (Figure 7l). Aurora A kinase might therefore be a downstream target of RHAMM/TPX2 in the nucleus. The results raise the new idea that RHAMM is essential for the activation of Aurora A kinase. We performed a flow cytometric analysis to assess the cell cycle progression of the RHAMM-knockdown cells in the presence of HA. In HA-added control cells, the percentage of the cells in the G2/M phase was 3.8±1.3% at 0 h and 47.9±1.0% at 48 h (Figure 7m). In the RHAMM-knockdown cells, this percentage was 2.6±0.45% at 0 h and 16.2±0.74% at 48 h (Figure 7m). The percentage of cells in the G2/M phase was considerably reduced in the RHAMM-knockdown cells compared with control cells at 48 h under HA-added conditions. These results indicated that RHAMM was involved in the accumulation of G2/M cells and has an important role in cell cycle progression in HCF cells. Taken together, the present results indicate that RHAMM/TPX2 interaction participates in activating Aurora A kinase and are consistent with the finding that RHAMM is required for the accumulation of cells in the M phase.
DISCUSSION
In this study, RHAMM was found to positively regulate ERK in cementifying fibroma cells. The signaling mediated by mitogen-activated protein kinases (MAPKs) has been shown to have critical roles in the regulation of cell growth and differentiation.34 Recent studies have raised the possibility that RHAMM could regulate MAPKs such as ERK, which control the expression of proteins relevant to cell proliferation and motility.21 However, the precise mechanism by which RHAMM responds to the intracellular signaling pathway in mammalian cells is not well understood. We found that RHAMM could associate with ERK and promote the growth of HCF cells. Our findings are consistent with prior research showing that RHAMM has a significant role in controlling MAPKs.
Several previous studies have demonstrated that RHAMM is able to mediate HA-induced signaling in several types of cells.19, 20 Consistent with previous findings, RHAMM was found to be involved in HA-dependent signaling in HCF cells. In the presence of HA, RHAMM could function as a downstream target of CD44 and positively control the growth of HCF cells through a mechanism involving ERK. Given that the HA-induced phosphorylation of Raf, MEK, and ERK was reduced by inhibition of CD44, the Raf–MEK–ERK pathway could be major target of CD44. It is noteworthy that RHAMM was expressed in the centrosome and mitotic spindle and was associated with TPX2 in the presence of HA. Although the mechanisms through which RHAMM moves into the nucleus remain unknown, RHAMM and TPX2 seem to have important roles in the progression of the cell cycle. Given that the accumulation of cells in G2/M phase was inhibited by the knockdown of RHAMM, the association of RHAMM with the centrosome might ensure proper mitotic events in the cell cycle. However, complete inhibition of the cell cycle at G2/M was not observed in the RHAMM-knockdown HCF cells. The spindle-assembly checkpoint ensures the fidelity of chromosomal segregation and has an important role in cell cycle progression in the M phase.35 Therefore, an alternate mechanism such as the spindle-assembly checkpoint may affect the regulation of the cell cycle in HCF cells. In vivo, HA is a widely distributed component of the extracellular matrix, which is an important regulator of cellular proliferation, differentiation, and survival.36 In malignant cells, RHAMM is also likely to induce tumor progression by driving centrosome amplification and mitotic-spindle aberrations under HA-abundant condition.37 These results indicate that the function of RHAMM might be affected by the tumor microenvironment, including the extracellular matrix. In addition, we found that the HA-induced phosphorylation of Aurora kinase was inhibited by the elimination of RHAMM. RHAMM was thought to be involved in the activation of Aurora A. At the metaphase spindle, TPX2 is required for not only spindle pole integrity but also the activation of Aurora A in mammalian cells.38 Consequently, it is proposed that TPX2/RHAMM interaction is essential for the cell cycle progression through the activation of Aurora A.
EGF family ligands take part in the progression of cellular proliferation, migration, and angiogenesis.39 Epiregulin is a novel EGF ligand.27 In HCF cells, epiregulin was highly expressed and could function through an extracellular autocrine loop involving the constitutively activated EGFR. Epiregulin was able to stimulate the Raf–MEK–ERK signaling pathway in this study. These results support the idea that the constitutively activated EGFR has a fundamental role in the growth mechanisms of HCF cells. In addition, we have identified that epiregulin-induced ERK phosphorylation was suppressed by the elimination of RHAMM. The results suggest that RHAMM can function as an activator of ERK downstream of EGFR and is essential for epiregulin-dependent cell proliferation.
Interestingly, our studies disclosed that HA evoked the phosphorylation of EGFR in HCF cells. Furthermore, HA-induced cell proliferation was repressed by an EGFR inhibitor. This inhibitory effect appears to be mediated by the inhibition of EGFR activation. It seems likely that the downregulation of HA-induced cell proliferation was mostly due to the blocking of CD44-induced enhancement of the EGFR phosphorylation. Hence, our results raised the hypothesis that EGFR could respond to CD44 activation and participate in the HA-induced signaling pathway. Furthermore, HA-induced EGFR phosphorylation could be suppressed by inhibition of CD44. Our results suggest that CD44-HA binding has a pivotal role in augmentation of the activation of EGFR. Recently, Bourguignon et al40 reported that HA provoked interaction between CD44 and EGFR in squamous cell carcinoma cells. This report supports the notion that CD44 can participate in the activation of EGFR in response to HA.
The HA-induced phosphorylation of Raf, MEK, and ERK was substantially attenuated by the EGFR inhibitor, which blocked tyrosine kinase activity of EGFR. These results raise the possibility that the HA-induced activation of MAPKs mainly depends on the EGFR. Based on these results, we propose that the EGFR has an essential role in the cell proliferation elicited by HA. Figure 8 schematically represents our conclusion that RHAMM functions as an activator of ERK, thought to be regulated by the epiregulin-induced activation of EGFR and the HA-induced activation of CD44/EGFR. Here, we report that RHAMM induces proliferative activity through a novel mechanism based on the CD44–EGFR. Moreover, it is novel finding that RHAMM has a pivotal role in activation of Aurora A in the nucleus. The EGFR–RHAMM/ERK signaling pathway has been implicated in growth and the cell cycle in HCF cells.
Proposed model of HA and epiregulin-induced signal transduction in HCF cells. HA-CD44 binding induces a Raf–MEK–ERK signal cascade via the activation of EGFR. RHAMM functions as an activator of ERK in EGFR downstream signaling. RHAMM could associate not only with ERK1/2, leading to tumor cell growth, but also with TPX2 in the nucleus in response to HA. At the same time, RHAMM localizes in the centrosomes and mitotic spindle and can function as an activator of Aurora A, leading to progression of the mitotic stage of the cell cycle. Moreover, epiregulin functions as a ligand of EGFR through extracellular autocrine loops. Epiregulin can activate Raf–MEK–ERK signaling via EGFR.
Cementifying fibroma usually consists of multiplied fibroblasts that produce extracelluar collagen fibers and osteoblastic cells on the surface of spherules-bone/cementum granules. HA is part of the extracellular matrix and likely to modulate tumor cell growth in the microenvironment of cementifying fibroma. Abundant biosynthesis of HA may positively regulate the growth of cementifying fibroma cells in a RHAMM–ERK-dependent manner.
In the in vivo study, we found that RHAMM protein expression could be observed in most of several cementifying fibroma cases examined (data not shown). Moreover, expression of RHAMM protein was detected in cells from the fibrous region of the tissues. These findings suggest that RHAMM might be expressed more generally in cementifying fibromas and have an important role in the growth of the cells. Additionally, we examined the mineralizing activity of HCF, but found the degree of activity to be lower than that of normal osteoblasts (Supplementary Figure 1b). The relationship between the RHAMM gene and differentiation including calcification especially in osteobasts or cementoblasts is not clear. Interestingly, ALP mRNA expression and ALP activity were enhanced by the RHAMM knockdown in HCF cells (Supplementary Figures 1c, 1d, and 1e). ALP activity is thought to be early osteogenic marker. These observations suggest a mechanism by which RHAMM/ERK functions as a negative regulator of mineralization in HCF cells. Recent studies showed that ERK-MAPK activity is associated with anti-osteogenesis in osteoblasts.41, 42, 43 A MAPK inhibitor as well as a dominant negative MEK1 enhanced the differentiation of osteoblasts.41, 42 These findings in vitro have significant implications in explaining the inhibitory effect on the differentiation of ERK-MAPK. Taken that the elimination of RHAMM represses the activation of the ERK, it is assumed that RHAMM/ERK can function as a negative regulator for osteogenic features of HCF cells. In addition to its role in cell proliferation, RHAMM could serve as an anti-osteogenic factor in the osteolytic stage in cementifying fibroma. RHAMM expression may be linked to characteristic features of cementifying fibroma, including cellular proliferation and differentiation. We conclude that RHAMM has a major role in the growth, cell cycle, and differentiation of cementifying fibroma cells.
References
Kramer IR, Pindborg JJ, Shear M . The WHO histological typing of odontogenic tumours. A commentary on the Second Edition. Cancer 1992;70:2988–2994.
Slootweg PJ . Osseous dysplasia, WHO classification of tumours. In: Barnes L, Eveson JW, Reichart, P et al. Pathology and Genetics, Head and Neck Tumours. IARC press: Lyon, 2005; 323.
Kudo Y, Hiraoka M, Kitagawa S, et al. Establishment of human cementifying fibroma cell lines by transfection with temperature-sensitive simian virus-40 T-antigen gene and hTERT gene. Bone 2002;30:712–717.
Wang C, Entwistle J, Hou G, et al. The characterization of a human RHAMM cDNA: conservation of the hyaluronan-binding domains. Gene 1996;174:299–306.
Hardwick C, Hoare K, Owens R, et al. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J Cell Biol 1992;117:1343–1350.
Assmann V, Jenkinson D, Marshall JF, et al. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J Cell Sci 1999;112:3943–3954.
Turley EA, Noble PW, Bourguignon LY, et al. Signaling properties of hyaluronan receptors. J Biol Chem 2002;277:4589–4592.
Bajorath J, Greenfield B, Munro SB, et al. Identification of CD44 residues important for hyaluronan binding and delineation of the binding site. J Biol Chem 1998;273:338–343.
Entwistle J, Hall CL, Turley EA, et al. HA receptors: regulators of signalling to the cytoskeleton. J Cell Biochem 1996;61:569–577.
Banerji S, Ni J, Wang SX, et al. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J Cell Biol 1999;144:789–801.
Laurent TC, Fraser JR . Hyaluronan. FASEB 1992;J6:2397–2404.
Kawasaki K, Ochi M, Uchio Y, et al. Hyaluronic acid enhances proliferation and chondroitin sulfate synthesis in cultured chondrocytes embedded in collagen gels. J Cell Physiol 1999;179:142–148.
Yoneda M, Suzuki S, Kimata K, et al. Hyaluronic acid associated with the surfaces of cultured fibroblasts is linked to a serum-derived 85-kDa protein. J Biol Chem 1990;265:5247–5257.
Mahadevan P, Larkins RG, Fraser JR, et al. Effect of prostaglandin E2 and hyaluronan on mesangial cell proliferation. A potential contribution to glomerular hypercellularity in diabetes. Diabetes 1996;45:44–50.
Goebeler M, Kaufmann D, Brocker EB, et al. Migration of highly aggressive melanoma cells on hyaluronic acid is associated with functional changes, increased turnover and shedding of CD44 receptors. J Cell Sci 1996;109:957–1964.
Kothapalli D, Flowers J, Xu T, et al. Differential activation of ERK and Rac mediates the proliferative and anti-proliferative effects of hyaluronan and CD44. J Biol Chem 2008;283:31823–31829.
Savani RC, Wang C, Yang B, et al. Migration of bovine aortic smooth muscle cells after wounding injury. The role of hyaluronan and RHAMM. J Clin Invest 1995;95:1158–1168.
Pilarski LM, Miszta H, Turley EA . Regulated expression of a receptor for hyaluronan-mediated motility on human thymocytes and T cells. J Immunol 1993;150:4292–4302.
Hall CL, Wang C, Lange LA, et al. Hyaluronan and the hyaluronan receptor RHAMM promote focal adhesion turnover and transient tyrosine kinase activity. J Cell Biol 1994;126:575–588.
Zang S, Chang MD, Zylka D, et al. The hyaluronan receptor RHAMM regulates extracellular regulated kinase. J Biol Chem 1998;273:11342–11348.
Hall CL, Yang B, Yang X, et al. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell 1995;82:19–26.
Shigeishi H, Fujimoto S, Hiraoka M, et al. Overexpression of the receptor for hyaluronan-mediated motility, correlates with expression of microtubule-associated protein in human oral squamous cell carcinomas. Int J Oncol 2009;34:1565–1571.
Crainie M, Belch AR, Mant MJ, et al. Overexpression of the receptor for hyaluronan-mediated motility (RHAMM) characterizes the malignant clone in multiple myeloma: identification of three distinct RHAMM variants. Blood 1999;93:1684–1696.
Teder P, Bergh J, Heldin P . Functional hyaluronan receptors are expressed on a squamous cell lung carcinoma cell line but not on other lung carcinoma cell lines. Cancer Res 1995;55:3908–3914.
Gust KM, Hofer MD, Perner SR, et al. RHAMM (CD168) is overexpressed at the protein level and may constitute an immunogenic antigen in advanced prostate cancer disease. Neoplasia 2009;11:956–963.
Maxwell CA, Keats JJ, Belch AR, et al. Receptor for hyaluronan-mediated motility correlates with centrosome abnormalities in multiple myeloma and maintains mitotic integrity. Cancer Res 2005;65:850–860.
Tsai MY, Wiese C, Cao K, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol 2003;5:242–248.
Toyoda H, Komurasaki T, Uchida D, et al. Epiregulin, a novel epidermal growth factor with mitogenic activity for rat primary hepatocytes. J Biol Chem 1995;270:7495–7500.
Kamata N, Fujimoto N, Tomonari M, et al. Immortalization of human dental papilla, dental pulp, periodontal ligament cells and gingival fibroblasts by telomerase reverse transcriptase. J Oral Pathol Med 2004;33:417–423.
Yamanouchi K, Satomura K, Gotoh Y, et al. Bone formation by transplanted human osteoblasts cultured within collagen sponge with dexamethasone in vitro. J Bone Miner Res 2001;16:857–867.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Method 2001;25:402–408.
Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 2002;30:e15.
Sleeman JP, Kondo K, Moll J, et al. Variant exons v6 and v7 together expand the repertoire of glycosaminoglycans bound by CD44. J Biol Chem 1997;12:31837–31844.
Roberts PJ, Der CJ . Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007;26:3291–3310.
Hartwell LH, Kastan MB . Cell cycle control and cancer. Science 1994;16:1821–1828.
Noble PW . Hyaluronan and its catabolic products in tissue injury and repair. Matrix Biol 2002;21:25–29.
Pujana MA, Han JD, Starita LM, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet 2007;39:1338–1349.
Kufer TA, Silljé HH, Körner R, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol 2002;158:617–623.
Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2–16.
Bourguignon LY, Gilad E, Brightman A, et al. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem 2006;281:14026–14040.
Nakayama K, Tamura Y, Suzawa M, et al. Receptor tyrosine kinases inhibit bone morphogenetic protein-Smad responsive promoter activity and differentiation of murine MC3T3-E1 osteoblast-like cells. J Bone Miner Res 2003;18:827–835.
Higuchi C, Myoui A, Hashimoto N, et al. Continuous inhibition of MAPK signaling promotes the early osteoblastic differentiation and mineralization of the extracellular matrix. J Bone Miner Res 2002;17:1785–1794.
Schindeler A, Little DG . Ras-MAPK signaling in osteogenic differentiation: friend or foe? J Bone Miner Res 2006;21:1331–1338.
Acknowledgements
This work was supported by a Grant-in-aid (No. 17791461) from the Japanese Ministry of Education, Culture, Sports, and Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Supplementary information
Rights and permissions
About this article
Cite this article
Hatano, H., Shigeishi, H., Kudo, Y. et al. RHAMM/ERK interaction induces proliferative activities of cementifying fibroma cells through a mechanism based on the CD44–EGFR. Lab Invest 91, 379–391 (2011). https://doi.org/10.1038/labinvest.2010.176
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2010.176
Keywords
This article is cited by
-
The importance of RHAMM in the normal brain and gliomas: physiological and pathological roles
British Journal of Cancer (2023)
-
Extracellular matrix and its therapeutic potential for cancer treatment
Signal Transduction and Targeted Therapy (2021)
-
Enrichment of in vivo transcription data from dietary intervention studies with in vitro data provides improved insight into gene regulation mechanisms in the intestinal mucosa
Genes & Nutrition (2017)
-
Low molecular weight hyaluronan induces migration of human choriocarcinoma JEG-3 cells mediated by RHAMM as well as by PI3K and MAPK pathways
Histochemistry and Cell Biology (2017)
-
CD44 and RHAMM are essential for rapid growth of bladder cancer driven by loss of Glycogen Debranching Enzyme (AGL)
BMC Cancer (2016)