Abstract
Breast cancer is a major cause of morbidity and mortality in women and its metastatic spread is the principal reason behind the fatal outcome. Metastasis-related research of breast cancer is however underdeveloped when compared with the abundant literature on primary tumors. We applied an unexplored approach comparing at high resolution the genomic profiles of primary tumors and synchronous axillary lymph node metastases from 13 patients with breast cancer. Overall, primary tumors displayed 20% higher number of aberrations than metastases. In all but two patients, we detected in total 157 statistically significant differences between primary lesions and matched metastases. We further observed differences that can be linked to metastatic disease and there was also an overlapping pattern of changes between different patients. Many of the differences described here have been previously linked to poor patient survival, suggesting that this is a viable approach toward finding biomarkers for disease progression and definition of new targets useful for development of anticancer drugs. Frequent genetic differences between primary tumors and metastases in breast cancer also question, at least to some extent, the role of primary tumors as a surrogate subject of study for the systemic disease.
Similar content being viewed by others
Introduction
Breast cancer is a major cause of morbidity and disease-related mortality in women. The hematogenic and lymphogenic metastatic spread of tumor cells is the principal reason behind the fatal outcome.1 Despite this, metastasis-related research of breast cancer is not well developed, as judged from the number of publications targeting metastatic disease compared with the literature focusing on primary tumors. Axillary lymph node (ALN) metastasis status is the major predictor of survival as well as one of the key parameters used in staging of the disease, which has implications in clinical decision-making. Patients with ALN negative disease have a better prognosis. However, a substantial fraction of patients with ALN metastases will not develop distant metastases to other organs. On the other hand, a subset of patients free from ALN metastases will develop distant metastases and frequently succumb to the disease.1, 2 This lack of strong correlation illustrates the heterogeneity of underlying mechanisms driving disease progression.
In recent years, the application of high-throughput technologies for molecular characterization of breast cancer has emerged as a new and promising method for molecular classification. Studies have focused on gene expression microarrays aiming at improvement of prognostic classification of the primary tumors and characterization of the major events involved in tumor progression in breast cancer.3, 4, 5, 6 This led to the delineation of several molecular subtypes of the disease that carry prognostic value. Array-based profiling of gene copy number, sometimes in combination with global or locus-specific expression analysis, has also made an impact on breast cancer research. Multiple genomic aberrations found in primary tumors have been linked to poor prognosis for the patient.7, 8, 9, 10 Primary tumors and matched ALN metastases from breast cancer patients have been studied by low-resolution genomic approaches for assessment of copy number changes, such as metaphase CGH, allelic imbalance and low-resolution array-CGH.11, 12, 13, 14 In one report using metaphase CGH,12 divergence between the primary tumors and ALN metastases was noted in several cases. In another recent report, genome-wide array-CGH with average resolution of 1 Mb per data point was used in the analysis of ALN metastases and primary tumors from 29 patients.14 Minor differences were noted in only three subjects.
We have applied a high-resolution DNA copy number analysis of matched primary breast tumors and ALN metastases to delineate biomarkers of disease progression. The motivation for this study stems from our previous analysis of genetic variation and genetic heterogeneity in somatic cells, both for normal and tumor-derived samples.15, 16, 17 One of our previous reports, surveying ∼1% of the human genome, suggested a frequent intratumoral (within the same primary tumor) heterogeneity of genetic aberrations in breast cancer.15 We therefore hypothesized that there likely exist differences between the genetic profiles of primary tumors and corresponding metastases from the same patient. Global genome copy number variation (CNV) analysis of 13 breast cancer patients revealed frequent genetic differences between matched primary breast tumors and ALN metastasis and these revealed previously characterized biomarkers of disease progression.
Materials and methods
Patient material
Samples from 13 primary breast tumors and their corresponding ALN metastases were obtained from female patients treated at the Oncology Centre, Bydgoszcz, Poland. Patient clinical data are shown in Table 1 . The patients studied here represent a fraction of >150 breast cancer patient samples that were consecutively collected. Availability of samples from primary tumor and matched synchronous metastasis was the only criterion for inclusion in this study and all samples fulfilling this criterion were used. This study has been approved by the Research Ethics Committee of the Oncology Centre, Bydgoszcz, Poland, and the IRB of the University of Alabama at Birmingham, AL, USA.
32K BAC array-CGH, data processing, statistical analysis and pathway mining
High molecular weight DNA was extracted using standard protocols. For each patient, we carried out four hybridizations (duplicates of primary tumor and metastasis) to 32K BAC arrays, according to a well-established scheme of dye-swap experiments as described earlier.15, 16, 17, 18 Image acquisition was performed using the GenePix 4000B scanner (Axon Instruments Inc, Union City, CA, USA). Analysis of hybridization intensity was carried out using the GenePixPro v6 image analysis software (Axon Instruments). Storage and analysis of raw data were performed using the Linnaeus Centre for Bioinformatics (LCB) platforms: BASE Laboratory Information Management System (see https://baselims.lcb.uu.se/) and Data WareHouse (see https://dw.lcb.uu.se/). Using filters provided by LCB platforms, we first removed all non-optimal array features, which included oversaturated (>5%) spots, spots with low signal-to-noise ratio (SNR<3) in either of the channels, or those flagged as bad, absent or not found in the GenePixPro program. To remove possible dye bias or spatial effects, we also normalized all data using a print-tip loess normalization method. We then applied a dye-swap validation filter, which removed clones with a standard deviation of >0.15 between standard and the dye-swap replicate experiment. Finally, the array-CGH profiles from duplicate experiments were merged. Scoring of regions displaying copy number aberrations was performed using commercial software Nexus-CGH (BioDiscovery, El Segundo, CA, USA), which uses the circular binary segmentation algorithm. The statistical significance of regions with differences within the same patient between primary tumor and metastasis was computed using a Bonferroni-corrected two-sample t-test as described earlier.16 The t-test compares values within the region of interest with those outside it. A Bonferroni correction is applied to resulting P-values to account for the bias introduced by choosing the most salient regions, among all possible regions, for testing. P-values are therefore multiplied by the number of possible contiguous regions on a chromosome, namely  , where ni is the number of probed loci on chromosome i. This conservative approach lends assurance that statistically significant regions reflect an underlying biological phenomenon and are not merely the result of measurement error and selection bias. Candidate breast cancer genes were selected on the basis of information available in public breast cancer databases: http://www.itb.cnr.it/breastcancer/ and http://www.breastcancerdatabase.org/ as well as within the Ingenuity Pathways Analysis, a commercial software for analyses of gene/protein functions and interactions.
, where ni is the number of probed loci on chromosome i. This conservative approach lends assurance that statistically significant regions reflect an underlying biological phenomenon and are not merely the result of measurement error and selection bias. Candidate breast cancer genes were selected on the basis of information available in public breast cancer databases: http://www.itb.cnr.it/breastcancer/ and http://www.breastcancerdatabase.org/ as well as within the Ingenuity Pathways Analysis, a commercial software for analyses of gene/protein functions and interactions.
Results
32K genomic profiling of primary tumors and matched ALN metastases
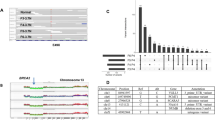
Primary and metastatic tumors from each patient were analyzed in four experiments with 32K BAC array covering the human genome; two hybridizations for primary tumor DNA and two hybridizations for metastasis, according to a dye-swap procedure. All hybridizations of cancer-derived DNA were carried out against blood DNA from a healthy female control, which was previously used in multiple studies.15, 16, 18 Results from two hybridizations for each primary tumor and from two hybridizations for each metastasis were merged and initially analyzed as separate experiments. For scoring and visualization of copy number changes, we applied the rank segmentation procedure using Nexus-CGH software. The detailed clinical and molecular description of the studied samples is shown in Table 1. A summary of the numbers of aberrations observed and the classification of the genomic profiles derived from separate analyses of primary tumors and metastases is described in Table 2 . We classified results from these individual array-CGH experiments of primary tumors and metastases using a previously reported scheme,9, 10 with minor modifications. Four subtypes of profiles were identified: (a) simplex; (b) complex; (c) spike/simplex; and (d) flat. The ‘simplex’ pattern was characterized by the presence of a few low copy number (up to four copies) genetic aberrations. In the ‘complex’ pattern, the majority of chromosomes were affected by the presence of multiple low or moderate copy number intertwined alterations. Moderate copy number was defined as the presence of four to six copies of a chromosomal segment. The ‘spike/simplex’ pattern was characterized by the presence of at least one distinct, highly amplified peak (higher than six copies) or clusters of such peaks restricted to a single chromosome arm on a background of a profile that could be classified as simplex. The ‘flat’ profile had no obvious changes (Table 2; Figure 1; Supplementary Figure 1a and b).
The examples of four classes of global genome CNV pattern comparing matched primary tumor and metastasis of patients 67, 23, 26 and 127 affected by breast cancer. The panel for each case represents a summary of four hybridizations using the 32K BAC array: two for primary tumor DNA and two for metastases. Each hybridization of cancer-derived DNA was carried out against DNA from peripheral blood of a normal healthy female control. Red and blue dots represent data from the merging of two hybridizations for metastasis and primary tumor, respectively. The X-axis denotes chromosome mapping and the Y-axis represents a straight fluorescent ratio from hybridization of tumor-derived DNA against a normal female control. The ‘no. spots’ value represents the sum of array features from merged duplicate hybridizations that passed the quality control criteria for scoring. Only spots that had a standard deviation of ⩽15% of ratios in regular and dye-swap hybridizations were subject to further statistical analysis (see Materials and Methods)., On the basis of statistically significant differences in the level of changes between profiles of primary tumors and metastases, a combination of bioinformatics and statistical tools resulted in the division of array-CGH profiles into four classes. For a detailed definition of the four classes, see Results. In panel displaying case ID 23, an enlarged view of chromosome 1 is shown with a reversed order of color layers for red (metastasis) and blue (primary tumor).
The total number of aberrations was higher in primary tumors than in metastases and the overall difference was ∼20%. The average number of aberrations per analyzed sample was 33 in primary tumors (range 6–83) and 27 for metastases (range 2–64). Overall, the gains were predominant over deletions, but this difference was mainly due to a higher number of gains in primary tumors. In metastases, the number of gains and deletions was similar (Table 2). We identified common regions of aberrations frequently present as gains at 1q, 8q, 11q, 16p, 17q and 20q as well as deletions at 1p, 8p, 11q, 16q and 17p and the majority of our observations were consistent with previous analyses of primary tumors from breast cancer patients.7, 9, 19 Supplementary Figure 2 shows the overall frequency plots of aberrations combined for all primary tumors versus all metastases. No statistically significant difference in frequency of aberrations between these two groups was detected using Fisher's exact test. This statistical analysis is different from tests of significance for specific genetic aberrations that differ between primary tumor and matched metastasis (see above Materials and Methods, below and Supplementary Table 1). Evident areas of highly amplified and often clustered peaks were seen in multiple patients (Figure 1; Supplementary Figure 1). These were often, but not always, present in both metastasis and primary tumor and were located at chromosome 1, 4, 8, 10, 11, 17, 19 and 20. One of the strongest amplified regions, located within 11q13.1–11q14.1 (64.47–78.3 Mb) is shown in detail in Figure 2. Mining of databases and literature suggested that this interval encompasses 16 candidate breast cancer-related genes (Table 3 ; Supplementary Table 2). The region within 17q12–q21.2 (33.76–35.83 Mb) encompassed at least 22 cancer-related genes and was also previously reported as having strong association between DNA copy number gain/amplification and increased gene expression.20, 21, 22
Two high-resolution images of the amplicon from 11q13.1–11q14.1 (64.47–78.3 Mb) in four patients. In two of these (IDs 22 and 67), the profiles of primary tumor (blue dots) and metastasis are highly correlated. In two cases (IDs 23 and 140), these amplicons are pronounced in metastases. The X-axis describes the position on chromosome 11 and the Y-axis represents a straight fluorescent ratio from hybridization of tumor-derived DNA against a normal female control. For example, a straight fluorescent ratio of five corresponds to ∼10 DNA copies at a given locus. The ‘no. spots’ value represents the sum of the displayed array features from merged duplicate hybridizations that passed the quality control criteria for scoring.
Four classes of combined aberration profiles for primary tumors versus metastases
Comparison of profiles for matched primary tumor and ALN metastasis revealed numerous differences in terms of presence or absence of aberrations between these samples. Alternatively, the difference between a primary tumor and metastasis was evident by the change in DNA copy number level: for example, a higher level of amplification in metastasis for an amplicon that was also present in the primary tumor. We determined the statistical significance of the differences between the primary tumor and corresponding metastasis using a Bonferroni-corrected two-sample t-test16 (Figure 1; Supplementary Figure 1 and Supplementary Table 1). This approach is inherently conservative and helps to assure that the identified regions represent actual CNV and not merely measurement noise. On the basis of these results, combined primary tumor/metastasis profiles were grouped into four classes. In Class I, profiles of primary tumor and metastasis were essentially indistinguishable. Class II patients had many differences in the number of aberrations and the magnitude of copy number change present in primary tumor and metastasis. Furthermore, these differences between the primary tumor and metastasis profiles in a single patient were ‘bidirectional,’ meaning that some chromosomal regions were presenting a change in primary tumor, whereas other regions displayed the reverse, with a particular change present only in metastasis. Overall, class II patients represent a highly complex picture, and this aberration class encompasses the majority of studied subjects. Class III was characterized by ‘unidirectional’ changes detectable in metastasis, which were not present in primary tumor. Finally, class IV is the opposite of class III, with significant aberrations present exclusively in primary tumor.
Figure 1 shows four whole genome profiles with combined analysis of primary tumor (blue dots) and metastasis (red dots), each graph representing one of the four above-described classes. Supplementary Figure 1 shows pairwise comparisons for all remaining cases. Supplementary Table 1 displays details of all regions scored as deviating between primary tumors and metastases. This table summarizes data from the analysis of 11 cases showing 157 statistically significant differences, with a majority of chromosomes being involved. Only two chromosomes (5 and 14) did not display any significant differences between the primary tumors and metastases. Considering the large number of regions with deviations, we applied an additional scoring criterion for selection of regions with most pronounced changes, by calculation of differences between values of average fluorescent ratio for primary tumor and metastasis for each region and selection of regions with >20% difference. Forty-four deviations that were identified in this way are highlighted by asterisks in Supplementary Tables 1 and 3.
Aberration class I is represented by two cases (IDs 22 and 67), which showed tumor-specific aberrations in analyses of primary tumors and metastases affecting many chromosomes with a few narrow and high amplification peaks present at chromosome 5, 11q and 17p. However, none of these differed significantly between primary tumor and metastasis (Figures 1 and 2; Supplementary Figures 1 and 3). Aberration class II constitutes the bulk of patients (7 cases). It is also the most heterogeneous group and encompasses the largest number of differences uncovered between primary tumors and metastases (Supplementary Table 1). Many of the differences were shared by cases within class II or across classes (Table 3). In particular, a few of these located at chromosomes 1, 4, 8 and 11 and represented as gains or amplifications were shared by up to four patients, and were characterized by a high level of difference between metastasis and primary tumor. The majority of these regions encompassed genes for which DNA amplification is strongly correlated with overexpression, and genes that have been previously shown as differentially expressed in metastasis relative to primary tumor20, 21, 22, 23, 24, 25, 26 (Table 3; Supplementary Table 2). For example, an aberrant region unique to class II at 6q15–6q16 (87.55–97.56 Mb) and exclusively deleted in metastasis of breast cancer contains candidate tumor suppressor genes described earlier for prostate cancer: PNRC1 and CASP8AP2.27 PNRC1 gene expression was previously found to be significantly reduced in breast cancer tissue relative to uninvolved margin,28 which may indicate its role as a tumor suppressor. A second region with deletion in metastasis at 11q11–11q12.1 (56.06–56.51 Mb) encompassed genes encoding olfactory receptors and this aberration was also observed in a previously reported profiling of primary breast cancers.10 In two additional regions located at 9q31.3–9q33.1 (113.74–121.9 Mb) and 10q21.1–10q21.2 (59.88–63.32 Mb), both gains and deletions in metastases were present (Table 3). We were unable to obtain data from the literature on the involvement of the genes present in the latter region in metastasis, whereas the former contained two candidate genes, DBC1 and DEC1, involved in the regulation of apoptosis.29, 30 Aberration class III is represented by only one patient (ID 26), and it is characterized by two broad segments of gain at 1q and at 16p. Both the differences between primary tumor and metastasis observed in this subject have also been seen in other patients from aberration classes II and IV (Table 3). Aberration class IV, represented by three cases (IDs 35, 127 and 131), has unique regions with aberrations on chromosome 20. In particular, region 20q11–q13 (29.47–47.34 Mb) encompasses 10 genes, the expression of which is strongly correlated with DNA amplification22 (Supplementary Table 2).
In at least four of the studied cases (IDs 23, 30, 140 and 26), we observed clear genetic differences that can be linked to metastatic breast cancer. For instance, patient 23 (class II) (Figures 1 and 2) displayed two high copy number gains on 8p (>8 copies) and 11q (>10 copies) that were more prominent in metastasis. Both aberrations have already been linked to poor survival of breast cancer patients.22, 31 Many of the aberrations differing between primary tumors and metastases occurred in several patients. Table 3 shows a summary of these selected regions and the relevant case ID. It should be noted, however, that a substantial number of these showed inconsistent patterns of change between primary tumor and metastasis. For instance, a certain chromosomal region could display a gain that was only present in metastasis for one patient, whereas the same chromosomal segment was instead amplified in a primary tumor in another subject. An illustrative example of such a contradictory pattern of change is shown in Figure 1 and Supplementary Figure 1 for patients 23, 26 and 62, involving a low copy number gain of the long arm of chromosome 1. Supplementary Table 2 describes the results from mining of databases and literature with regard to candidate genes that are located in the chromosomal regions that deviate between primary tumors and metastases and occur in at least two cases.
Discussion
Our study is the first that compares CNV profiles for primary tumors and metastases from patients with breast cancer samples globally and with high resolution. On the basis of large genomic clones with an average resolution of 50–100 kb per data point,16, 17, 18, 32 we applied a well established, thoroughly validated array covering ∼99% of the human genome. The average resolution of our analysis is more than an order of magnitude higher than that used in the recent array-CGH analysis comparing ALN metastases and primary tumors.14 This is the most likely reason for discrepancies in results between the two studies. The experimental approach we took is also sensitive and robust, which is well illustrated by a strong correlation between findings from our work and previously published reports of breast cancers focusing on primary tumors.7, 8, 9, 10 Our methodology is also insensitive to DNA CNV that might be present in normal DNA of the studied patients.
On the basis of the analysis of global gene expression profiles, there is a debate in the field whether primary tumors from breast cancer patients and matched ALN metastases are different or not, as contradictory results have been published.23, 24, 25, 26, 33, 34 Many factors can be responsible for this incongruence: both related to the still poorly understood biology and a well-recognized heterogeneity of the disease, low numbers of studied patients as well as factors related to the differences in the methodology. Our results from the analysis of DNA copy number actually support both conclusions. The aberration class I cases (IDs 22 and 67) show essentially indistinguishable profiles and cases such as these should also display very similar profiles of gene expression in the primary tumor sample and corresponding metastasis. Although we have not investigated gene expression, the literature supports this assumption. Previous publications describing the parallel analysis of gene copy number changes and mRNA expression in primary tumors of breast cancer have reported a strong correlation between the findings derived from both types of analysis.7, 20, 21, 22, 31 On the other hand, the aberration class I cases are the minority in our study and all the remaining patients showed more or less pronounced differences in gene DNA copy number profiles. This would presumably result in dissimilarity in the levels of transcripts produced from regions affected by gains/amplifications or deletions. The aberration class I patients also raise questions regarding the timing and dynamics behind the process of ALN metastases and the number of cells derived from primary tumors that colonize a lymph node. The highly similar pattern of genetic aberrations in these matched samples may be a reflection of a large number of cells that colonized the lymph node and this metastasis has since then not increased considerably in cell number and therefore did not develop a different genomic profile.
We observed that the total number of aberrations was higher in primary tumors than in metastases and this may seem surprising at first glance. However, this finding likely reflects the heterogeneity of different clones of cells having different genetic profiles that are present within a primary tumor, which is related to the multistage process of cancer development. Our results and others from primary breast tumors suggest frequent intratumoral heterogeneity of genetic aberrations12, 15, 35, 36, 37, 38 implying a coexistence, within the same mass of primary tumor, of different cell subpopulations with different genetic profiles. Metastasis is likely established by a minority of cells from the primary tumor, which migrate to a single distant site. One can therefore envisage a bottleneck ‘purifying’ effect for aberrations that a metastasis will contain at the time of its formation, which then may or may not develop into a profile differing from primary tumor by acquisition of additional genetic changes. It should also be emphasized that the above-mentioned previous studies of intratumoral heterogeneity of breast cancer require reanalysis using methodologies that fulfill the current standards of resolution for global genome analysis. The ‘flat’ pattern of array-CGH profiles that was characteristic for two metastases (aberration class IV, IDs 35 and 127) also deserves a comment. This could simply be the result of heavy (>90%) non-cancerous cell contamination. This explanation is, however, less probable as the tissues were assessed for tumor cell content by a pathologist. The existence of such flat genomic profiles with no chromosomal alterations in primary tumors has been reported in the past.8, 10, 22 Furthermore, such findings are compatible with a newer model for parallel metastasis development. It has been shown that the early disseminated tumor cells, the potential metastatic progenitors, are genetically significantly less aberrant than the matched primary tumors.39, 40 The above issue requires further analysis in a considerably larger cohort of patients.
Although we studied a limited number of paired samples, we noticed numerous differences in amplifications/gains and deletions between matched samples, suggesting the differential activation of oncogenes and inactivation of tumor suppressor genes, respectively, in primary tumors versus metastases. Aberrations that are detected in metastases and are not present in primary tumors, as well as changes that are more pronounced in metastases (eg higher level of amplification) compared with primary tumors, represent candidate biomarkers for disease progression and merit further study to delineate the specific gene (or genes) that may be involved. The above statement is based on a reasonable assumption that ALN metastases represent an expansion of a more aggressive clone of cells derived from a primary tumor. We observed many clear-cut differences that can be linked to progression of the disease. For instance, the DNA of case ID 23 displayed two high copy number gains on 8p and 11q that were more prominent in metastasis. The series of amplicons on 11q (64.47–78.3 Mb) that has been mentioned above involves many well-characterized cancer-related genes. Both 11q and 8p changes have been previously linked to poor survival of breast cancer patients.22, 41, 42 This and other examples of the known progression-related changes observed in our study suggest that our reasoning and approach toward finding biomarkers for breast cancer progression is correct. By analogy, the genetic aberrations described here that are acquired in metastases and that are not yet linked to poor patient survival, for example 1q (148–152 Mb), 6q (87–97 Mb), 10q (43–45 Mb) and 11p (56.06–56.51 Mb), might also be important and should be studied further. Moreover, we have recently completed the experimental phase of an ovarian cancer-related project. We performed a similar analysis comparing matched primary tumors and metastases to omentum, and the results also indicate frequent differences in DNA CNV profiles between such sample pairs (Poplawski et al, in preparation). There also seems to be an overlap between progression-related genetic changes seen in a breast cancer study and those from ovarian cancer. In summary, the most valuable implication of this report is that our approach has the potential to enhance the characterization of specific genes that are linked to breast cancer progression, which forms the basis for the development of new anticancer drugs. The frequent genetic differences between primary tumors and metastases also question, at least to some extent, the role of primary tumors as a surrogate subject of study for the systemic disease, when the development of efficient new molecular therapy is considered.43, 44 Our study calls for an extension, using a considerably larger number of patients who have also been followed for a longer time. In an ideal scenario, it would be important to evaluate many samples from each patient; that is, several for the same primary tumor, several for different ALN metastases as well as samples derived from distant metastases.
References
Polyak K : Breast cancer: origins and evolution. J Clin Invest 2007; 117: 3155–3163.
Eccles S, Paon L, Sleeman J : Lymphatic metastasis in breast cancer: importance and new insights into cellular and molecular mechanisms. Clin Exp Metastasis 2007; 24: 619–636.
Bild AH, Potti A, Nevins JR : Linking oncogenic pathways with therapeutic opportunities. Nat Rev 2006; 6: 735–741.
Driouch K, Landemaine T, Sin S, Wang S, Lidereau R : Gene arrays for diagnosis, prognosis and treatment of breast cancer metastasis. Clin Exp Metastasis 2007; 24: 575–585.
Sotiriou C, Piccart MJ : Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev 2007; 7: 545–553.
Stadler ZK, Come SE : Review of gene-expression profiling and its clinical use in breast cancer. Crit Rev Oncol Hematol 2009; 69:1–11.
Haverty PM, Fridlyand J, Li L et al: High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer 2008; 47: 530–542.
Chin SF, Wang Y, Thorne NP et al: Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene 2007; 26: 1959–1970.
Fridlyand J, Snijders AM, Ylstra B et al: Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer 2006; 6: 96.
Hicks J, Krasnitz A, Lakshmi B et al: Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 2006; 16: 1465–1479.
Kuukasjarvi T, Karhu R, Tanner M et al: Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 1997; 57: 1597–1604.
Torres L, Ribeiro FR, Pandis N, Andersen JA, Heim S, Teixeira MR : Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res Treat 2007; 102: 143–155.
Becker TE, Ellsworth RE, Deyarmin B et al: The genomic heritage of lymph node metastases: implications for clinical management of patients with breast cancer. Ann Surg Oncol 2008; 15: 1056–1063.
Li J, Gromov P, Gromova I et al: Omics-based profiling of carcinoma of the breast and matched regional lymph node metastasis. Proteomics 2008; 8: 5038–5052.
Benetkiewicz M, Piotrowski A, Diaz De Stahl T et al: Chromosome 22 array-CGH profiling of breast cancer delimited minimal common regions of genomic imbalances and revealed frequent intra-tumoral genetic heterogeneity. Int J Oncol 2006; 29: 935–945.
Bruder CE, Piotrowski A, Gijsbers AA et al: Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet 2008; 82: 763–771.
Piotrowski A, Bruder CE, Andersson R et al: Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat 2008; 29: 1118–1124.
Diaz de Stahl T, Sandgren J, Piotrowski A et al: Profiling of copy number variations (CNVs) in healthy individuals from three ethnic groups using a human genome 32 K BAC-clone-based array. Hum Mutat 2008; 29: 398–408.
Hicks DG, Yoder BJ, Short S et al: Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease-free survival in subsets of breast cancer patients. Clin Cancer Res 2006; 12: 6702–6708.
Arriola E, Marchio C, Tan DS et al: Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest 2008; 88: 491–503.
Pollack JR, Sorlie T, Perou CM et al: Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA 2002; 99: 12963–12968.
Chin SF, Teschendorff AE, Marioni JC et al: High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol 2007; 8: R215.
Hao X, Sun B, Hu L et al: Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer 2004; 100: 1110–1122.
Suzuki M, Tarin D : Gene expression profiling of human lymph node metastases and matched primary breast carcinomas: clinical implications. Mol Oncol 2007; 1: 172–180.
Feng Y, Sun B, Li X et al: Differentially expressed genes between primary cancer and paired lymph node metastases predict clinical outcome of node-positive breast cancer patients. Breast Cancer Res Treat 2007; 103: 319–329.
Vecchi M, Confalonieri S, Nuciforo P et al: Breast cancer metastases are molecularly distinct from their primary tumors. Oncogene 2008; 27: 2148–2158.
Lapointe J, Li C, Giacomini CP et al: Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res 2007; 67: 8504–8510.
Zhou D, Chen B, Ye JJ, Chen S : A novel crosstalk mechanism between nuclear receptor-mediated and growth factor/Ras-mediated pathways through PNRC-Grb2 interaction. Oncogene 2004; 23: 5394–5404.
Ehata S, Hanyu A, Hayashi M et al: Transforming growth factor-beta promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res 2007; 67: 9694–9703.
Kim JE, Chen J, Lou Z : DBC1 is a negative regulator of SIRT1. Nature 2008; 451: 583–586.
Gelsi-Boyer V, Orsetti B, Cervera N et al: Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res 2005; 3: 655–667.
Nord H, Hartmann C, Andersson R et al: Characterization of novel and complex genomic aberrations in glioblastoma using a 32K BAC array. Neuro Oncol 2009; E-pub ahead of print 12 June 2009.
Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van’t Veer LJ : Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA 2003; 100: 15901–15905.
Weigelt B, Wessels LF, Bosma AJ et al: No common denominator for breast cancer lymph node metastasis. Br J Cancer 2005; 93: 924–932.
Bonsing BA, Devilee P, Cleton-Jansen AM, Kuipers-Dijkshoorn N, Fleuren GJ, Cornelisse CJ : Evidence for limited molecular genetic heterogeneity as defined by allelotyping and clonal analysis in nine metastatic breast carcinomas. Cancer Res 1993; 53: 3804–3811.
Fujii H, Marsh C, Cairns P, Sidransky D, Gabrielson E : Genetic divergence in the clonal evolution of breast cancer. Cancer Res 1996; 56: 1493–1497.
Shen CY, Yu JC, Lo YL et al: Genome-wide search for loss of heterozygosity using laser capture microdissected tissue of breast carcinoma: an implication for mutator phenotype and breast cancer pathogenesis. Cancer Res 2000; 60: 3884–3892.
Mangia A, Chiarappa P, Tommasi S et al: Genetic heterogeneity by comparative genomic hybridization in BRCAx breast cancers. Cancer Genet Cytogenet 2008; 182: 75–83.
Schardt JA, Meyer M, Hartmann CH et al: Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell 2005; 8: 227–239.
Schmidt-Kittler O, Ragg T, Daskalakis A et al: From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA 2003; 100: 7737–7742.
Albertson DG : Gene amplification in cancer. Trends Genet 2006; 22: 447–455.
Paterson AL, Pole JC, Blood KA et al: Co-amplification of 8p12 and 11q13 in breast cancers is not the result of a single genomic event. Genes Chromosomes Cancer 2007; 46: 427–439.
Stoecklein NH, Hosch SB, Bezler M et al: Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell 2008; 13: 441–453.
Gow CH, Chang YL, Hsu YC et al: Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol 2009; 20: 696–702.
Acknowledgements
We thank Drs Eva Tiensuu Janson and Devin Absher for a critical review of the manuscript. This study was supported by the University of Alabama at Birmingham, the Swedish Cancer Foundation and the Swedish Children Cancer Fund. SWE was supported by NIH grant T32 HL072757.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Popławski, A., Jankowski, M., Erickson, S. et al. Frequent genetic differences between matched primary and metastatic breast cancer provide an approach to identification of biomarkers for disease progression. Eur J Hum Genet 18, 560–568 (2010). https://doi.org/10.1038/ejhg.2009.230
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.230
Keywords
This article is cited by
-
The genomic regulation of metastatic dormancy
Cancer and Metastasis Reviews (2023)
-
Evolution of early phase clinical trials in oncology
Journal of Molecular Medicine (2018)
-
P-cadherin: a useful biomarker for axillary-based breast cancer decisions in the clinical practice
Modern Pathology (2017)
-
In stage II/III lymph node-positive breast cancer patients less than 55 years of age, keratin 8 expression in lymph node metastases but not in the primary tumour is an indicator of better survival
Virchows Archiv (2015)
-
Biological resonance for cancer metastasis, a new hypothesis based on comparisons between primary cancers and metastases
Cancer Microenvironment (2013)