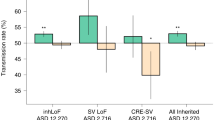
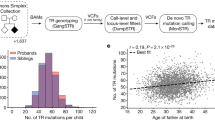
Abstract
The identification of the genetic components of autism spectrum disorders (ASDs) has advanced rapidly in recent years, particularly with the demonstration of de novo mutations as an important source of causality. We review these developments in light of genetic models for ASDs. We consider the number of genetic loci that underlie ASDs and the relative contributions from different mutational classes, and we discuss possible mechanisms by which these mutations might lead to dysfunction. We update the two-class risk genetic model for autism, especially in regard to children with high intelligence quotients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fombonne, E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 65, 591–598 (2009).
Muhle, R., Trentacoste, S. V. & Rapin, I. The genetics of autism. Pediatrics 113, e472–e486 (2004).
Newschaffer, C. J. et al. The epidemiology of autism spectrum disorders. Annu. Rev. Publ. Health 28, 235–258 (2007).
Power, R. A. et al. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse versus their unaffected siblings. JAMA Psychiatry 70, 22–30 (2013)
Ku, C. S. et al. A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol. Psychiatry 18, 141–153 (2013).
Bailey, A. et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psych Med. 25, 63–77 (1995).
Rosenberg, R. E. et al. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch. Pediatr. Adolesc. Med. 163, 907–914 (2009).
Constantino, J. N., Zhang, Y., Frazier, T., Abbacchi, A. M. & Law, P. Sibling recurrence and the genetic epidemiology of autism. Am. J. Psychiatry 167, 1349–1356 (2010).
Amir, R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet. 23, 185–188 (1999).
Fu, Y. H. et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67, 1047–1058 (1991).
Verkerk, A. J. et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 (1991).
Klei, L. et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol. Autism 3, 9 (2012).
Lee, S. H. et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genet. 45, 984–994 (2013).
Devlin, B. & Scherer, S. W. Genetic architecture in autism spectrum disorder. Curr. Opin. Genet. Dev. 22, 229–237 (2012).
Marshall, C. R. et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82, 477–488 (2008).
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007).
Veltman, J. A. & Brunner, H. G. De novo mutations in human genetic disease. Nature Rev. Genet. 13, 565–575 (2012).
Ozonoff, S. et al. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics 128, e488–e495 (2011).
Jorde, L. B. et al. Complex segregation analysis of autism. Am. J. Hum. Genet. 49, 932–938 (1991).
Risch, N. et al. A genomic screen of autism: evidence for a multilocus etiology. Am. J. Hum. Genet. 65, 493–507 (1999).
Zhao, X. et al. A unified genetic theory for sporadic and inherited autism. Proc. Natl Acad. Sci. USA 104, 12831–12836 (2007).
Fischbach, G. D. & Lord, C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68, 192–195 (2010).
Geschwind, D. H. et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 69, 463–466 (2001).
Robinson, E. B., Lichtenstein, P., Anckarsater, H., Happe, F. & Ronald, A. Examining and interpreting the female protective effect against autistic behavior. Proc. Natl Acad. Sci. USA 110, 5258–5262 (2013).
Iafrate, A. J. et al. Detection of large-scale variation in the human genome. Nature Genet. 36, 949–951 (2004).
Sebat, J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004).
Levy, D. et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70, 886–897 (2011).
Pinto, D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–372 (2010).
Sanders, S. J. et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams Syndrome region, are strongly associated with autism. Neuron 70, 863–885 (2011).
Lee, Y. H. et al. Reducing system noise in copy number data using principal components of self–self hybridizations. Proc. Natl Acad. Sci. USA 109, E103–E110 (2012).
Bamshad, M. J. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature Rev. Genet. 12, 745–755 (2011).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nature Genet. 39, 1522–1527 (2007).
Ng, S. B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Iossifov, I. et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012).
Neale, B. M. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012).
O'Roak, B. J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012).
Sanders, S. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012).
Lundstrom, S. et al. Trajectories leading to autism spectrum disorders are affected by paternal age: findings from two nationally representative twin studies. J. Child Psychol. Psychiatry 51, 850–856 (2010).
Waller, D. K. et al. The population-based prevalence of achondroplasia and thanatophoric dysplasia in selected regions of the US. Am. J. Med. Genet. A 146A, 2385–2389 (2008).
Zammit, S. et al. Paternal age and risk for schizophrenia. Br. J. Psychiatry 183, 405–408 (2003).
Kong, A. et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature 488, 471–475 (2012).
Gilman, S. R. et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 70, 898–907 (2011).
O'Roak, B. J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Auerbach, B. D., Osterweil, E. K. & Bear, M. F. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 480, 63–68 (2011).
Bear, M. F., Huber, K. M. & Warren, S. T. The mGluR theory of fragile X mental retardation. Trends Neurol. 27, 370–377 (2004).
Gregg, C., Zhang, J., Butler, J. E., Haig, D. & Dulac, C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329, 682–685 (2010).
DeVeale, B., van der Kooy, D. & Babak, T. Critical evaluation of imprinted gene expression by RNA-seq: a new perspective. PLoS Genet. 8, e1002600 (2012).
Barlow, D. P. Genomic imprinting: a mammalian epigenetic discovery model. Ann. Rev. Genet. 45, 379–403 (2011).
Cantor, R. M. et al. Replication of autism linkage: fine-mapping peak at 17q21. Am. J. Hum. Genet. 76, 1050–1056 (2005).
Chen, G. K., Kono, N., Geschwind, D. H. & Cantor, R. M. Quantitative trait locus analysis of nonverbal communication in autism spectrum disorder. Mol. Psych 11, 214–220 (2006).
McCauley, J. L. et al. Genome-wide and ordered-subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Med. Genet. 6, 1 (2005).
Ylisaukko-oja, T. et al. Search for autism loci by combined analysis of Autism Genetic Resource Exchange and Finnish families. Ann. Neurol. 59, 145–155 (2006).
Anney, R. et al. A genomewide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 19, 4072–4082 (2010).
Wang, K. et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459, 528–533 (2009).
Weiss, L. A., Arking, D. E., Daly, M. J. & Chakravarti, A. A genome-wide linkage and association scan reveals novel loci for autism. Nature 461, 802–808 (2009).
Murdoch, J. D. & State, M. W. Recent developments in the genetics of autism spectrum disorders. Curr. Opin. Genet. Dev. 23, 310–315 (2013).
Davies, G. et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry 16, 996–1005 (2011).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nature Genet. 42, 565–569 (2010).
Lim, E. T. et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron 77, 235–242 (2013).
Girirajan, S. & Eichler, E. E. Phenotypic variability and genetic susceptibility to genomic disorders. Hum. Mol. Genet. 19, R176–R187 (2010).
Hallmayer, J. et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 68, 1095–1102 (2011).
Talkowski, M. E. et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149, 525–537 (2012).
Evrony, G. D. et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–496 (2012).
Hancks, D. C. & Kazazian, H. H. Jr. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203 (2012).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Lindhurst, M. J. et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. New Engl. J. Med. 365, 611–619 (2011).
Feldman, I., Rzhetsky, A. & Vitkup, D. Network properties of genes harboring inherited disease mutations. Proc. Natl Acad. Sci. USA 105, 4323–4328 (2008).
Blake, J. A., Bult, C. J., Kadin, J. A., Richardson, J. E. & Eppig, J. T. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 39, D842–D848 (2011).
Bayes, A. et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nature Neurosci. 14, 19–21 (2011).
Voineagu, I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 25, 25–29 (2000).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Coincident mutations
-
Mutations in both alleles at a given locus.
- Comparative genomic hybridization
-
(CGH). A microarray-based technique for identifying large deletions or duplications in the genome.
- Concordance
-
The probability that multiple siblings are affected given that one of them is already known to be affected.
- Copy number variants
-
(CNVs). Large deletions or duplications that either alter the number of copies of genes or disrupt the function of genes.
- De novo mutations
-
New mutations that arise either in the parental germ line or somatically.
- Dosage sensitivity
-
A defining feature of phenotypes that result from heterozygous mutation.
- Gender bias
-
The phenomenon whereby four times as many males are affected by autism spectrum disorders compared with females, with a male:female ratio of nearly 6:1 among individuals who are diagnosed as being high functioning.
- High-risk families
-
Families that contain a highly penetrant segregating risk allele for autism spectrum disorders.
- Insertions and deletions
-
(Indels). Small insertions or deletions in the genome that are generally <10 bp.
- Loss-of-function mutations
-
In the context of this article, events that result in a nonsense allele or that change the reading frame.
- Low-risk families
-
Families that do not contain a segregating risk allele for autism spectrum disorders (ASDs) and that are only at risk of ASDs in cases of de novo mutation.
- Monoallelic
-
Pertaining to the expression of only one allele at a given locus.
- Multiplex families
-
Families with multiple affected children.
- Neuroplasticity
-
The dynamic state of the brain, which enables it to respond to changes in environment and development.
- Penetrant
-
Pertaining to the probability that an individual with a given mutation will be affected by the corresponding condition.
- Recurrence
-
Independent mutational 'hits' within a given gene in unrelated individuals.
- Sibling risk
-
The probability that a sibling of an affected child will also be affected.
- Simplex families
-
Families with only one affected child; all other children (if any) of these families are unaffected.
- Transmitted
-
Inheritance of a mutant allele from a parent, who may be phenotypically normal owing to gender bias.
- Trios
-
Family units that consist of both parents and one child in each unit.
Rights and permissions
About this article
Cite this article
Ronemus, M., Iossifov, I., Levy, D. et al. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet 15, 133–141 (2014). https://doi.org/10.1038/nrg3585
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3585
This article is cited by
-
Severity of Autism Spectrum Disorder Symptoms Associated with de novo Variants and Pregnancy-Induced Hypertension
Journal of Autism and Developmental Disorders (2024)
-
CGH Findings in Children with Complex and Essential Autistic Spectrum Disorder
Journal of Autism and Developmental Disorders (2023)
-
Examining Diagnostic Trends and Gender Differences in the ADOS-II
Journal of Autism and Developmental Disorders (2023)
-
Whole Genome Analysis of Dizygotic Twins With Autism Reveals Prevalent Transposon Insertion Within Neuronal Regulatory Elements: Potential Implications for Disease Etiology and Clinical Assessment
Journal of Autism and Developmental Disorders (2023)
-
Altered Metabolic Characteristics in Plasma of Young Boys with Autism Spectrum Disorder
Journal of Autism and Developmental Disorders (2022)