Abstract
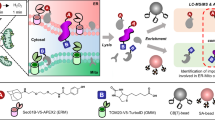
Activity-based protein profiling (ABPP) is a functional proteomics technique for directly monitoring the expression of active enzymes in cell extracts and living cells. The technique relies on irreversible inhibitors equipped with reactive groups (warheads) that covalently attach to the active site of enzymes and fluorescent or affinity tags for imaging and purification purposes, respectively. Here, a high-throughput and robust protocol for high-resolution quantitative activity-based proteasome profiling is described. We use both panreactive and subunit-specific fluorescent activity-based probes (ABPs) to quantify the proteasome activity in living cells, in the presence or absence of the potent proteasome inhibitor bortezomib. Active proteasome subunits from cell lysates are affinity-purified via a biotinylated ABP. Purification from live cells involves a two-step ABP approach using a reagent with a cell-permeable azide-warhead and postlysis installation of biotin. By means of liquid chromatography–mass spectrometry (LC-MS)-based proteomics, we can accurately identify the enriched proteins and the active site peptides of the enzymes, and relatively quantify all the proteasome activities in one experiment. The fluorescence ABPP protocols takes 2–3 d, and approximately 8–10 d are needed to complete the entire protocol.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rock, K.L. & Goldberg, A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17, 739–779 (1999).
Kloetzel, P.M. & Ossendorp, F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 16, 76–81 (2004).
Kane, R.C., Bross, P.F., Farrell, A.T. & Pazdur, R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist 8, 508–513 (2003).
Jung, T., Catalgol, B. & Grune, T. The proteasomal system. Mol. Aspects Med. 30, 191–296 (2009).
Marques, A.J., Palanimurugan, R., Matias, A.C., Ramos, P.C. & Dohmen, R.J. Catalytic mechanism and assembly of the proteasome. Chem. Rev. 109, 1509–1536 (2009).
Borissenko, L. & Groll, M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 107, 687–717 (2007).
Murata, S. et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 (2007).
Verdoes, M., Florea, B.I., van der Marel, G.A. & Overkleeft, H.S. Chemical tools to study the proteasome. Eur. J. Org. Chem. 20, 3301–3313 (2009).
Kisselev, A.F. & Goldberg, A.L. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 398, 364–378 (2005).
Li, N., Overkleeft, H.S. & Florea, B.I. Activity-based protein profiling: an enabling technology in chemical biology research. Curr. Opin. Chem. Biol. 16, 227–233 (2012).
Fenteany, G. et al. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268, 726–731 (1995).
Meng, L. et al. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96, 10403–10411 (1999).
Bogyo, M. et al. Covalent modification of the active site threonine of proteasomal β-subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA 94, 6629–6634 (1997).
Kessler, B.M. et al. Extended peptide-based inhibitors efficiently target the proteasome and reveal overlapping specificities of the catalytic β-subunits. Chem. Biol. 8, 913–929 (2001).
Berkers, C.R. et al. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat. Methods 2, 357–362 (2005).
Verdoes, M. et al. A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem. Biol. 13, 1217–1226 (2006).
Florea, B.I. et al. Activity-based profiling reveals reactivity of the murine thymoproteasome-specific subunit beta5t. Chem. Biol. 17, 795–801 (2010).
Verdoes, M. et al. A panel of subunit-selective activity-based proteasome probes. Org. Biomol. Chem. 8, 2719–2727 (2010).
Kraus, M. et al. Activity patterns of proteasome subunits reflect bortezomib sensitivity of hematologic malignancies and are variable in primary human leukemia cells. Leukemia 21, 84–92 (2007).
Ruckrich, T. et al. Characterization of the ubiquitin-proteasome system in bortezomib-adapted cells. Leukemia 23, 1098–1105 (2009).
Saxon, E. & Bertozzi, C. Cell surface engineering by a modified Staudinger reaction. Science 287, 2007–2017 (2000).
Ovaa, H. et al. Chemistry in living cells: detection of active proteasomes by a two-step labeling strategy. Angew. Chem. Int. Ed. 42, 3626–3629 (2003).
Willems, L.I. et al. Triple bioorthogonal ligation strategy for simultaneous labeling of multiple enzymatic activities. Angew. Chem. Int. Ed. 51, 4431–4434 (2012).
Boersema, P.J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Raijmakers, R. et al. Target profiling of a small library of phosphodiesterase 5 (PDE5) inhibitors using chemical proteomics. Chem.Med.Chem. 5, 1927–1936 (2010).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 (2009).
Kisselev, A.F., Songyang, Z. & Goldberg, A.L. Why does threonine, and not serine, function as the active site nucleophile in proteasomes? J. Biol. Chem. 275, 14831–14837 (2000).
Singh, J., Petter, R.C., Baillie, T.A. & Whitty, A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 (2011).
Muchamuel, T. et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 15, 781–787 (2009).
Kallemeijn, W.W. et al. Novel activity-based probes for broad-spectrum profiling of retaining β-exoglucosidases in situ and in vivo. Angew. Chem. Int. Ed. 51, 12529–12533 (2012).
Chang, J.T. et al. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity 34, 492–504 (2011).
Adibekian, A. et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat. Chem. Biol. 7, 469–478 (2011).
Weerapana, E., Speers, A.E. & Cravatt, B.F. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2, 1414–1425 (2007).
Lin, G. et al. Inhibitors selective for mycobacterial versus human proteasomes. Nature 461, 621–626 (2009).
Gu, C. et al. Proteasome activity profiling: a simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J. 62, 160–170 (2010).
van der Linden, W.A. et al. Two-step bioorthogonal activity-based proteasome profiling using copper-free click reagents: a comparative study. Bioorg. Med. Chem. 20, 662–666 (2012).
Wessel, D. & Flugge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J.V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Olsen, J.V. et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteomics 4, 2010–2021 (2005).
Acknowledgements
We are grateful to the Netherlands Genomics Initiative, the Netherlands Proteomics Center and the Netherlands Organization for Scientific Research (NWO) for funding this work.
Author information
Authors and Affiliations
Contributions
N.L. carried out the cell culture (treatment), affinity purification, SDS-PAGE imaging work and prepared the first draft of the manuscript and subsequent modifications. C.-L.K. and G.P. carried out the cell culture (treatment), affinity purification and SDS-PAGE imaging work. M.V., L.I.W., W.A.v.d.L. and M.R. refined the design and synthesis of the chemical tools used in this work. H.v.d.E., E.v.G. and J.G. performed LC-MS analysis and data processing. G.P.v.W., H.S.O. and B.I.F. supervised the work and were involved in manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Li, N., Kuo, CL., Paniagua, G. et al. Relative quantification of proteasome activity by activity-based protein profiling and LC-MS/MS. Nat Protoc 8, 1155–1168 (2013). https://doi.org/10.1038/nprot.2013.065
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.065
This article is cited by
-
Carfilzomib resistance due to ABCB1/MDR1 overexpression is overcome by nelfinavir and lopinavir in multiple myeloma
Leukemia (2018)
-
C3a triggers formation of sub-retinal pigment epithelium deposits via the ubiquitin proteasome pathway
Scientific Reports (2018)
-
Mapping in vivo target interaction profiles of covalent inhibitors using chemical proteomics with label-free quantification
Nature Protocols (2018)
-
Activity-based probes for functional interrogation of retaining β-glucuronidases
Nature Chemical Biology (2017)
-
A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function
Nature Structural & Molecular Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.


