Abstract
In the previous studies, the populations in Tibet exhibited a complicated genetic structure, indicating that those populations might be the admixture of East Asian and South/Central Asian populations, or have a North Asian origin. However, there have not been sufficient genetic data to support this hypothesis. In this study, we analyzed 15 autosomal polymorphic tetranucleotide short tandem repeat loci (D5S818, FGA, D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51) for three populations from Tibet, namely, Deng/Mishmi (n=114), Qamdo Tibetan (n=78) and Lhasa Tibetan (n=101). The total number of observed alleles and the average heterozygosity for all samples were 394 and 0.7574, respectively. Analysis of molecular variance and estimated GST (0.0198) for these allele frequency data suggested the genetic divergence among Tibetan populations was significant. Furthermore, our new allele frequency data for 13 loci were compared with those of 41 world populations previously reported. Results from phylogenetic and multidimensional scaling analyses indicated that: (1) the Deng in Tibet has unique genetic characteristics different from the Tibetans; (2) populations living in the Himalayas area (Deng, Luoba/Adi) composed of a distinct cluster and are closely related to each other than to any other ethnic groups in East Asia; (3) the Tibetans are most similar to the North Asians. This genetic structure is consistent with the geographical barriers and linguistic classifications.
Similar content being viewed by others
Introduction
Tibet, a land full of mystery in the southwest of China, bordering on India, Nepal, Bhutan, Burma and so on, has many minorities other than the Tibetans, most of which have seldom been studied genetically. The most distinctive one of the minorities in Tibet, Deng, also known as Mishmi (means uncivilized people), is a unique population in the Himalayas in Southeast Tibet (29°N, 96°E) surrounded by the Tibetans and Luoba/Adi people. The languages of Deng (two dialects: Darang and Geman) were classified as North Assam branch of Sino-Tibetan family Tibeto-Burman subfamily (http://www.ethnologue.com), together with the Luoba languages. However, with a total population of around 28 000 (2001 census) but only 1000 in the area controlled by China, the ethnic affiliation of Deng is yet to be determined officially in China, although Deng people claim to be descendents of Luoba (an official minority of China).1 Neither of the Deng or Luoba people have any feeling of identity with the Tibetans, the dominant population of Tibet whose languages form a Himalayish Tibetan branch of Tibeto-Burman subfamily, and are divided into three distinctive major branches (Figure 1): Weizang (Central Tibetan in Lhasa, Rikaze, Shannan and so on), Amdo (northern Tibetan in Qinghai, Gansu and Aba prefecture of Sichuan), Khams (Eastern Tibetan in Ganzi of Sichuan, Deqing of Yunnan and Qamdo of Tibet)2 and four minor branches. Previous studies using classical genetic traits,3 autosomal microsatellite markers4, 5 and mitochondrial DNA6 suggest a North Asian origin of Tibetans, while evidences from the Y chromosomal Alu insertion (YAP) marker reveal much more intricate stories for the origin of Tibetan peoples.7, 8, 9, 10 Not like the Tibetan studies, genetic study on Deng population is totally absent in literature to date, and nothing is known about their origin. Therefore, a genetic study of Deng and comparative analyses with the relevant populations, including Luoba, Tibetans and other East Asians, may shed light on the origin of the Himalayan unique populations.
Geographical locations of the 13 Tibetan-Himalayan population samples and distributions of the Tibetan ethnic branches. Reference populations out of the Tibetan Plateau are marked with triangles. Four minor branches (Choni, Ladak, Sherpa-Dzongkha and Kenkha) are also shown in the map beside the three major branches.
Short tandem repeats (STRs), also known as microsatellites, are most widely used to elucidate human population histories11, 12, 13, 14 and population structures.15 Moreover, the STR loci are especially valuable for the study of genetic relationships of closely related populations.16, 17, 18, 19 In this study, we applied the autosomal STR variation analysis to three population samples from Tibet to explore the peopling of the Himalayas.
Materials and methods
Population samples
The population samples collected and analyzed in this study include Deng (n=114) from Zayü County of Nyingchi Prefecture, and Tibetans from Lhasa Prefecture (n=101) and Qamdo Prefecture (n=78). All volunteers gave their informed content before their participation of the study. As we have mentioned, languages of Deng are within the North Assam branch of Tibeto-Burman and the Tibetan languages within the Himalayan branch. The Lhasa Tibetan is Central Tibetan (Weizang), and the Qamdo Tibetan is Eastern Tibetan (Khams or Kangba). The geographical locations and other general information of these samples are in Figure 1 and Table 1. To obtain a global picture of the genetic affiliation of the Tibetan Plateau populations to the populations of interest, data on 41 populations were compiled from literature20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52 (Table 1), including fourteen Tibeto-Burman populations, five Han Chinese populations, seven Altaic populations, five Tai-Kadai populations, two Hmong-Mien populations, one Austronesian population, three Austro-Asiatic populations and two Indo-Iranian populations in addition to the data on Europeans and Africans. Considering a possible connection between Deng and Luoba/Adi linguistically, all Luoba samples in the area controlled by China and Adi samples in the area controlled by India that have been studied are included in the subsequent analyses.
STR genotyping
Whole-blood samples were collected in EDTA vacutainer tubes by venipuncture from unrelated healthy indigenous individuals of Tibet. Ancestry of the samples was ascertained for three generations back in order to define autochthony. Genomic DNA was extracted by the standard phenol–chloroform procedure53 or the Chelex-100 protocol.54 For each sample, 15 most widely used forensic loci were amplified simultaneously using AmpFl STR Identifier PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA) at the D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818 and FGA STR loci. PCR amplifications were performed using 1.0–2.5 ng DNA amount in a final volume of 12 μl. DNA was amplified in a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer, Applied Biosystems). Amplified STR fragments were analyzed with an ABI PRISM 3100 Avant DNA Analyzer (Applied Biosystems Division/Perkin-Elmer). An internal size standard (GeneScan 500 LIZ, Perkin-Elmer, Applied Biosystems) was included. We also sequenced some samples for each locus to confirm the repeat number. Genotyping of each sample was analyzed using GeneScan 3.7 and Genotyper 3.7 software (Applied Biosystems, Foster City, CA, USA) by comparison with supplied allelic ladders. For some samples releasing peaks of abnormal shapes during genotyping, we chose to redo the genotyping or resequence to prevent spurious amplification. Allelic designations followed the recommendations of the DNA Commission of the International Society for Forensic Haemogenetics (DNA recommendations, 1994).
Statistical analyses
The allele frequencies were calculated by a single-gene counting method. Arlequin software version 3.1 was used to obtain observed and expected heterozygosity.55, 56 Tests for Hardy–Weinberg equilibrium were performed using a likelihood ratio test57 and an exact test58 to prevent miscalling STR genotypes or biased sampling. Locus-by-locus hierarchal analysis of molecular variance is carried out using Arlequin 3.1 software.
The GST values and Ht were estimated using DISPAN (http://mep.bio.psu/downlods/dispan.zip). The genetic distances were calculated from the allele frequency data at all the 13 STR loci (excluding D2S1338 and D19S433) by DA59 distance with the NJBAFD (http://iubio.bio.indiana.edu/soft/molbio/evolve/njbafd/), and θW (FST) distance60 with the PHYLIP 3.65c.61 The phylogenetic trees were constructed by the Neighbor-joining (NJ) method62 using the MEGA v4.063 and the allele frequency data, at 13 STR loci, of 44 populations by combining three populations typed in this study and 41 other populations obtained from the literature. It should be noted that two STR loci (D2S1338 and D19S433) were removed from the phylogenetic analysis because these two loci were not typed for many reference populations from literature. Bootstrap values were obtained based on 1000 replications.
The multidimensional scaling (MDS) analysis, based on pairwise DA distance values calculated at 13 STR loci in 44 populations, was performed using the SPSS 15.0 software package (SPSS, Chicago, IL, USA).
Results
Diversity of Deng and other Himalayan populations
Fifteen STR markers were typed in three populations sampled from Tibet (Deng, Lhasa Tibetan and Qamdo Tibetan) and their allele frequencies along with a number of genetic and polymorphic parameters of interest are provided in Supplementary Table 1. Deviation from Hardy–Weinberg equilibrium was tested for all possible loci by two methods: likelihood ratio test and exact test, respectively. No significant deviation was observed after Bonferroni correction for either test, indicating that our samples well represent the populations and most probably no miscalling of the STR allele happened.
To assess the diversity of Deng in comparison with other Tibetan-Himalayan (T-H) populations, the data of 15 STRs of other 10 T-H population samples (Figure 1) were added to our three population samples in the subsequent analyses. The total numbers of alleles were 127 (number of unique allele=8), 132 (1) and 135 (1) in Deng, Lhasa Tibetan and Qamdo Tibetan, respectively. The proportion of unique alleles varies from 0.74% (in Qamdo Tibetan) to 7.34% (in Adi Pasi1) in 13 T-H populations, and that of Deng is 6.30%. Unique alleles in the Tibetans are much fewer than the North Assam populations. The average heterozygosity values ranged from 0.7248 (Deng) to 0.7868 (Bhutanese) among 13 T-H populations, and that of Deng is the lowest (Table 2).
Phylogenetic analyses and genetic structure
A phylogenetic tree based on DA distances among 44 populations (Supplementary Table 2) was reconstructed by using the NJ method and shown in Figure 2. Deng, Luoba and the other six Adi populations first clustered with bootstrap value 67, indicating a close genetic relationship among these North Assam populations, all on the south of the Himalayas. The Tibetan populations also clustered with the North Assam populations with bootstrap value 67, forming a monophyletic structure. Other branches of the phylogeny are less reliable, given their small bootstrap values, although the population samples clustered well by the geographical distributions. Typing more loci will help to confirm the genetic relationships between these ethnic groups.
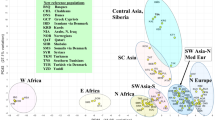
The results of MDS analysis using pairwise DA distance between populations demonstrated the genetic relationships among populations (Figure 3). Again, Luoba and six Adi populations form a cluster. However, Deng becomes an outlier of the cluster including all eight North Assam populations reported. The Tibetans are also close to each other. Interestingly, most of the Altaic samples (T2-T7 in Figure 3) from North Asia are closest to the Tibetans, indicating the close relationship between the Tibetans and North Asians, which has been hypothesized by many previous studies.3, 4, 5, 6
Multidimensional scaling (MDS) plot of 44 populations transformed from DA genetic distances using 13 autosomal STRs. Note: Codes are the same as those in Table 1.
Genetic differentiation between Deng and other Himalayan populations
Result from single analysis is usually unreliable. Here we tried to use multiple analyses to assess the population relationship. The GST values among populations, as a measure of genetic differentiation, were calculated as shown in Table 3. The GST value averaged over 15 loci were 0.0225, 0.0266 and 0.0333 among the seven Adi populations, eight North Assam populations (Adi and Deng) and 13 T-H populations, respectively, showing a trend of increased GST as more populations were included in the analysis. The GST value between Deng and seven Adi populations pooled (0.0144) was almost the same as that between Deng and five Tibetan populations pooled (0.0146), indicating almost the same genetic distance of the Deng from the Tibetans or the Adi, which was also reflected in the MDS plot of Figure 3. However, the GST value among the Tibetans (0.0075) was only half of that between the Tibetans and Deng (0.0146), and the GST value among the Adi populations (0.0225) was even double of that between Adi and Deng (0.0144), indicating that the Tibetans were all similar to each other, whereas North Assam populations were all quite different genetically. This distance pattern is also the same as that of the MDS plot, showing that Deng is a significant outlier of the Tibetans but not so distinct from the North Assam group to be excluded, as the North Assam populations are all quite different from each other. Therefore, Deng can only be grouped into the Adi/Luoba cluster.
The GST value among the four populations from Tibet (Deng, Lhasa Tibetan, Qamdo Tibetan and Luoba) was 0.0181, larger than the value among the Tibetans, Deng-Adi or Deng-Tibetans, but less than that among the seven Adi or eight North Assam. The four populations are all located in the present Tibet Autonomous Region, and therefore the relatively low GST value among these four populations may indicate the recent gene flows among them during the time of the Tibet Autonomous Region. The FST values among these four populations from Tibet were also estimated using analysis of molecular variance and were also shown in Table 3. The FST value, presented as a percentage of variation among populations, was 2.00% averaged over 15 loci, and was >3.65% at both TPOX and D13S317. The variances among the populations at locus TPOX was more significant than that of the other loci by analysis of molecular variance, indicating that TPOX is most variable among the 15 loci. The GST value averaged at TPOX was 0.0161 in the seven Adi populations, but about half of the values in the eight North Assam populations (0.0342), showing that this locus responds mostly for the difference between Adi and Deng. The Ht value averaged is 0.7792 at 15 loci in 13 populations, and is the lowest (0.6203) at TPOX.
The allele frequency distribution of Deng also show the characters different from other North Assam, for example, the frequencies of allele 10 at locus TH01 and allele 13 (CSF1PO) were 17.11 and 15% in Deng, while 0–1.2% and 3.5–8.6% in other seven Adi populations. The frequency of allele 11 at TPOX is 9.21% in Deng, and is 25–32.26% in Adi (Supplementary Table 1).
Discussion
Genetic segregation of the populations in Tibet
The aim of this study was to analyze the substructures of the populations residing in Tibet, and to examine the genetic relationship of the Tibetans and other Himalayan populations by using a set of autosomal markers. To achieve this aim, we analyzed genetic diversity of 293 unrelated individuals including Deng, Lhasa Tibetan and Qamdo Tibetan at 15 most commonly used autosomal STRs. These STR loci exhibited high diversity and were useful for the elucidation of population history and the genetic diversity among neighbor subpopulations. For technical considerations, these sites are easy for genotyping and scoring, and therefore, can be widely used to describe the population genetic feature.
In this study, our phylogenetic analyses of the population samples revealed that the populations in Tibet are quite similar to each other and different from the other East Asians and even far from the South Asians genetically (Figures 2 and 3), which is similar to the results of other studies.5, 9, 64, 65 Tibetan populations live mainly in the Tibetan plateau that located on the north of the Himalayas. The Himalayas harbors most of the highest peaks of the world and forms a natural barrier between the Tibetan plateau and the Indian subcontinent. These unique geographical features of the Tibetan landscape may have contributed to the genetic variety. However, we did see the close relationship between the Tibetans and the North Asians in this study, supporting the North Asian origin of the Tibetans suggested by many studies.3, 4, 5, 6
The long history of isolation in this plateau resulted in the T-H populations’ unique genetic structure. The history of T-H populations may be quite old.9 Archeological findings have revealed late Paleolithic inhabitation of the Tibetan plateau, dating the initial entry of modern humans to approximately 25–30 thousand years ago (KYA).66 However, the discovery of Neolithic sites,67 genetic data9, 68 and linguistic studies69 indicate populating of the plateau during the Neolithic period. Tibeto-Burman speakers are the major inhabitants of the Himalayas and the Tibetan Plateau. They occupy the territories of present Bhutan, Myanmar, Nepal, Assam and Tibet. This linguistic subfamily also extended into the eastern part of Southeast Asia.9, 65, 70 Ethnologically, the Tibeto-Burman subfamily corresponds to ‘Di-Qiang’ groups. According to history records, Di-Qiang tribes of northwestern China had migrated southward around 3 KYA, admixing with native residents on arrival.9, 10, 64, 65, 71 Su and coworkers suggested that the Bodic (on the north of the Himalayas) and Baric (on the south of the Himalayas) branches72 of the Tibeto-Burman subfamily populated Tibet and Nepal around 5–6 KYA.
Genetic structure of Deng population
Deng is a relatively small population in the Himalayas, with little contact with the people outside. Therefore, they were believed to be quite isolated and different to the neighboring populations genetically. However, little genetic studies have been conducted on this population and little was known about their origin. In this study, we found that STR allele distribution patterns exhibit considerable variation between Deng and other 12 neighboring populations. Deng showed somewhat lower range of total alleles (127) and higher of unique alleles (8) compared with the other 12 populations. The average observed heterozygosity of Deng was 0.7248, and was the lowest in 13 T-H populations, and the GST values of North Assam populations including Deng are higher than that excluding Deng (Table 3), reflecting Deng population's considerable isolation and inbreeding. These results were similar to those of the other isolated populations previously reported.13, 73, 74, 75 Lower gene diversity of Deng (0.7218) further accentuated the effect of the inbreeding among them. Therefore, we indicated that Deng might have developed from very few founders, and been isolated from the neighboring Adi and Tibetans for quite long time. However, compared with the results of the Andamanese73 by the same analyses, the time isolated of Deng should be much shorter. Judging from the phylogenetic tree and the MDS plot, Deng is closer to the Adi populations than to the Tibetans. Maybe, Adi/Luoba and Deng people have most recent common ancestors, supporting the claim of Deng people to be officially identified as Luoba.
Genetic relationship of Deng to other Himalayan or Asian population
Using DA genetic distances estimated from STR loci to measure the relationships among the populations is well accepted.76 In this paper, we applied DA distance to STR data of our population samples and 41 reference populations from literature including Adi/Luoba, Tibetans, Han Chinese, Indians, Japanese, Koreans and other populations (Table 1). In the NJ tree transformed from the DA distances (Figure 2), the main clusters of the tree were associated to the linguistic families and geographical distributions. Most of the bootstrap values were moderate to high, whereas some were quite low, such as those within Han Chinese populations, southwest minorities of China and Northeast Asians. These low values indicated that the 15 loci we used did not have high enough resolution for the structure within very similar population groups; thus more loci should be typed. However, for our T-H populations, these markers resulted in robust enough phylogenetic structure. The T-H populations clustered tightly, indicating gene flows or shared ancestors between these two groups. Similar to the close relationship between Deng and Adi revealed by the NJ tree, Deng was in the North Assam cluster and closest to the Luoba in the tree. Therefore, the NJ tree clearly illustrated that Deng was different from the Tibetans and close to the Adi/Luoba people, and the T-H populations were genetically quite far from the other East Asians, and even far from the Indians. MDS plot also showed a similar pattern with Deng being closest to the Adi cluster, and then the Tibetan cluster, and very far from the Indians. The genetic effects of the geographical barrier and ethnic segregation between Sino-Tibetan populations and Indians were pronounced, consistent with the results of Krithika et al.77
The genetic difference between North Assam populations and the East Asians shown by MDS plot might have resulted from either the gene flow from the South Asians to the North Assam populations or the genetic drift of these populations in the Himalayas. However, the Indians were even farther from the North Assam cluster in the MDS plot and NJ tree, suggesting no detectable gene flow from South Asians. Therefore, the deviation of the North Assam populations from the other East Asians was most probably resulted from the genetic drift. Deng and Adi/Luoba populations are all small and isolated in the valleys on the south of the Himalayas, and have a long history of inhabitancy in this area, living on hunting and gathering. The hard lifestyles had made the populations increase very slowly, or stop increasing, and sometimes even reduce, resulting in genetic drift.
The origin of Tibetans is widely debated. Previous genetic studies using classic markers,78 Y chromosome single-nucleotide polymorphism (SNP) and Y-STR,5 and mitochondrial DNA6 had depicted that Tibetans were clustered along with Northeast Asian group including Koreans, Japanese and Mongolians, thereby suggesting a North Asian origin. However, another report using Y-chromosome biallelic markers argued for the peopling of Tibet, Nepal and Bhutan by East Asians from the upper Yellow River region in China.9, 65, 68 Other studies suggested that the high frequency of the Y Alu insertion (YAP) in the Tibetan population signals a significant genetic contribution from Central Asia.8 Our result of NJ tree showed that Tibetan populations formed a distinctive cluster in the range of East Asians, not close to the Northeast or Southeast Asians but populations located closely beside the Tibetan Plateau; for example, Salar, Bai and Drung. However, the MDS plot showed that the Tibetans were quite close to the Altaic populations from North Asia, consistent with some of the previous studies.
In this study, we demonstrated the unique genetic structures of the Tibetan and Himalayan populations by analyzing the autosomal STR data. Further study using Y-STR, Y-SNP and mitochondrial DNA markers will be necessary to reconstruct more authentic history of the peopling of Tibet.
References
Huang, G. X. Ethnic Recognition in China (The Ethnic Publishing House, Beijing, 1995).
Guan, D. S. Grand Sight of the Ethnic Cultures in China, Tibetans, Monba, and Luoba, 1st edn (Encyclopedia of China Publishing House, Beijing, China, 1995).
Du, R., Xiao, C. & Cavalli-Sforza, L. L. Genetic distances between Chinese populations calculated on gene frequencies of 38 loci. Sci. China C Life Sci. 40, 613–621 (1997).
Starikovskaya, E. B., Sukernik, R. I., Derbeneva, O. A., Volodko, N. V., Ruiz-Pesini, E., Torroni, A. et al. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann. Hum. Genet. 69, 67–89 (2005).
Gayden, T., Cadenas, A. M., Regueiro, M., Singh, N. B., Zhivotovsky, L. A., Underhill, P. A. et al. The Himalayas as a directional barrier to gene flow. Am. J. Hum. Genet. 80, 884–894 (2007).
Torroni, A., Miller, J. A., Moore, L. G., Zamudio, S., Zhuang, J., Droma, T. et al. Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am. J. Phys. Anthropol. 93, 189–199 (1994).
Hammer, M. F., Spurdle, A. B., Karafet, T., Bonner, M. R., Wood, E. T., Novelletto, A. et al. The geographic distribution of human Y chromosome variation. Genetics 145, 787–805 (1997).
Qian, Y., Qian, B., Su, B., Yu, J., Ke, Y., Chu, Z. et al. Multiple origins of Tibetan Y chromosomes. Hum. Genet. 106, 453–454 (2000).
Su, B., Xiao, C., Deka, R., Seielstad, M. T., Kangwanpong, D., Xiao, J. et al. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum. Genet. 107, 582–590 (2000).
Shi, H., Zhong, H., Peng, Y., Dong, Y. L., Qi, X. B., Zhang, F. et al. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations. BMC Biol. 6, 45 (2008).
Jorde, L. B., Rogers, A. R., Bamshad, M., Watkins, W. S., Krakowiak, P., Sung, S. et al. Microsatellite diversity and the demographic history of modern humans. Proc. Natl Acad. Sci. USA 94, 3100–3103 (1997).
Zhivotovsky, L. A., Underhill, P. A., Cinnioglu, C., Kayser, M., Morar, B., Kivisild, T. et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population divergence time. Am. J. Hum. Genet. 74, 50–61 (2004).
Mansoor, A., Mazhar, K., Khaliq, S., Hameed, A., Rehman, S., Siddiqi, S. et al. Investigation of the Greek ancestry of populations from northern Pakistan. Hum. Genet. 114, 484–490 (2004).
Li, S. L., Yamamoto, T., Yoshimoto, T., Uchihi, R., Mizutani, M., Kurimoto, Y. et al. Phylogenetic relationship of the populations within and around Japan using 105 short tandem repeat polymorphic loci. Hum. Genet. 118, 695–707 (2006).
Rowold, D. J. & Herrera, R. J. On human STR sub-population structure. Forensic Sci. Int. 151, 59–69 (2005).
Chu, J. Y., Huang, W., Kuang, S. Q., Wang, J. M., Xu, J. J., Chu, Z. T. et al. Genetic relationship of populations in China. Proc. Natl Acad. Sci. USA 95, 11763–11768 (1998).
Rosenberg, N. A., Pritchard, J. K., Weber, J. L., Cann, H. W., Kidd, K. K., Zhivotovsky, L. A. et al. Genetic structure of human populations. Science 298, 2381–2385 (2002).
Ayub, Q., Mansoor, A., Ismail, M., Khaliq, S., Mohyuddin, A., Hameed, A. et al. Reconstruction of human evolutionary tree using polymorphic autosomal microsatellites. Am.J. Phys. Anthropol. 122, 259–268 (2003).
Zhang, F., Su, B., Zhang, Y. P. & Jin, L. Genetic studies of human diversity in East Asia. Phil. Trans. R. Soc. B. 362, 987–995 (2007).
Yan, J., Shen, C., Li, Y., Yu, X., Xiong, X., Mu, H. et al. Genetics analysis of 15 STR loci on Chinese Tibetan in Qinghai province. Forensic Sci. Int. 169, e3–e6 (2007).
Kraaijenbrink, T., van Driem, G. L., Karma, T. G. & de Knijff, P. Allele frequency distribution for 21 autosomal STR loci in Bhutan. Forensic Sci. Int. 170, 68–72 (2007).
Mansoor, A., Mazhar, K., Khaliq, S., Hameed, A., Rehman, S., Siddiqi, S. et al. Allele frequencies for 15 STR loci in Tibetan populations from Nepal. Forensic Sci. Int. 169, 234–238 (2007).
Kang, L. L. & Li, S. B. Allele frequencies of 15 STR loci of Luoba ethnic group in Tibet (Southwestern China). Forensic Sci. Int. 155, 219–221 (2005).
Krithika, S., Trivedi, R., Kashyap, V. K. & Vasulu, T. S. Allele frequency distribution at 15 autosomal STR loci Panggi, Komkar and Padam sub tribes of Adi, a Tibeto-Burman speaking population of Arunachal Pradesh, India. Legal Med. 9, 210–217 (2007).
Krithika, S., Trivedi, R., Kashyap, V. K. & Vasulu, T. S. Genetic diversity at 15 microsatelite loci among the Adi Pasi population of Adi tribal cluster in Arunachal Pradesh, India. Legal Med. 7, 306–310 (2005).
Lai, J. H., Chen, T., Feng, J. D., Zheng, H. B. & Li, S. B. Genetic polymorphism of fifteen short tandem repeat loci in Chinese Drung. Chin. J. Med. Genet. 19, 502–504 (2002).
Lai, J. H., Zhang, B. H., Zhu, B. F. & Li, S. B. STR polymorphism in Bai minority in Yunnan province. J. Xi’an Med. Univ. 23, 242–245 (2002).
Lai, Y., Wang, C. H., Qin, J., Jing, Q., Liu, J. X. & Xu, B. Y. Polymorphism investigation on 15 STR loci of Yunnan Lahu population. J. Kunming Med. Col. 3, 5–9 (2007).
Zhu, B. F., Shen, C. M., Wu, Q. J. & Deng, Y. J. Population data of 15 STR loci of Chinese Yi ethnic group. Legal Med. 10, 220–224 (2008).
Hu, S., Liu, J., Cai, K. & Lu, X. Genetics polymorphism of 15 STR loci in Chaoshan Han in China. Chin. J. Legal Med. 5, 287–289 (2004).
Wang, Z., Yu, R., Wang, F., Li, X. & Jin, T. Genetic polymorphisms of 15 STR loci in Han population from Shaanxi (NW China). Forensic Sci. Int. 147, 89–91 (2005).
Fan, X., Ding, J., Liang, Y., Xiao, B., Zhou, Y. & Liu, J. Fifteen short tandem repeat polymorphisms in Han race of North China. Chin. J. Med. Genet. 23, 227–229 (2006).
Liu, Y., Huo, Z. Y., Tang, H., Wang, J., Jiao, Z., Ma, W. et al. Frequency data for 15 STR loci and forensic use in a Beijing-Han population. Int. Congr. Ser. 1239, 267–270 (2003).
Hu, S. P., Wu, D. Q., Xu, X. H., Liu, J. W. & Li, B. Genetic profile of 15 STR loci in the Min Nan population in Southeast China. Forensic Sci. Int. 152, 263–265 (2005).
Zhu, B. F., Wang, Z., Wu, Q., Sedike, M. E., Zhu, J., Huang, P. et al. Genetic analysis of 15 STR loci of Chinese Uygur ethnic population. J. Forensic Sci. 50, 1235–1236 (2005).
Zhu, J., Shen, C., Ma, Y., He, Y., Zhao, J., Li, X. et al. Genetic polymorphisms of 15 STR in Chinese Salar ethnic minority group. Forensic Sci. Int. 173, 210–213 (2007).
Hashiyada, M., Itakura, Y., Nagashima, T., Nata, M. & Funayama, M. Polymorphism of 17 STRs by multiplex analysis in Japanese population. Forensic Sci. Int. 133, 250–253 (2003).
Kim, Y., Hwang, J., Kim, Y., Lee, S., Chung, N., Goh, H. et al. Allele frequencies of 15 STR loci using AmpF/STR Identifiler kit in a Korean population. Forensic Sci. Int. 136, 92–95 (2003).
Huang, Y., Gu, M., Wang, J., Du, Q., Li, X., Liu, M. et al. A genetic study of 15 STR loci in Chinese E Wen-ke population. J. Forensic Med. 20, 162–164 (2004).
Wang, J., Zhang, W., Huang, Y., Li, X., Gu, M. & Du, Q. A genetic study of 15 STR loci in Chinese Olunchun population. Forensic Sci. Technol. 5, 13–15 (2004).
Gu, M. B., Li, X. P., Du, Q. X., Wang, J., Huang, Y. L., Liu, M. et al. Polymorphism investigation on 15 STR loci of Inner-Mongolia Daur population. J. Forensic Med. 19, 166–167 (2004).
Yang, D., Liu, C. & Peng, R. B. Genetic study of 15 loci in southern China Han population and Li population. J. Forensic Med. 18, 207–210 (2002).
Yang, D., Liu, C., Liu, C. H., Wang, X. B., Zhu, S. J. & Li, F. H. Genetic polymorphic studies on 15 loci of 3 population in Guangxi province. J. Forensic Med. 17, 24–26 (2002).
Xu, L., Li, S. F., Deng, Q. Y., Zhou, L. N., Gong, J. C. & Xu, R. Polymorphism of fifteen tandem repeat loci in Maonan minority of Guangxi. Chin. J. Med. Genet. 24, 197–200 (2007).
Wang, P., Zhu, S. J., Liu, C., Li, Y., Li, F. G. & Liu, C. H. Genetic polymorphic studies on 15 loci of three ethnic groups in Guangxi province. J. Forensic Med. 18, 299–330 (2003).
Liu, C., Yang, D., Yang, C. H. & Zhu, S. J. Genetic studies of 15 STR loci in Guangxi Miao population. J. Forensic Med. 19, 204–206 (2003).
Othman, M. I., Seah, L. H., Panneerchelvam, S. & Nor, N. M. Allele frequencies for the PowerPlex 16 STR loci in Javanese population in Malaysia. J. Forensic Sci. 49, 190–191 (2004).
Sahoo, S. & Kashyap, V. K. Genetic variation at 15 autosomal microsatellite loci in the three highly endogamous tribal populations of Orissa, India. Forensic Sci. Int. 130, 189–193 (2002).
Shimada, I., Brinkmann, B., Tuyen, N. Q. & Hohoff, C. Allele frequency data for 16 STR loci in the Vietnamese population. Int. J. Legal Med. 116, 246–248 (2002).
Gaikwad, S. & Kashyap, V. K. Polymorphism at fifteen hypervariable microsatellite loci in four populations of Maharashtra, India. Forensic Sci. Int. 126, 267–271 (2002).
Camacho, M. V., Benito, C. & Figueiras, A. M. Allele frequencies of the 15 STR loci included in the AmpF/STR® identifilerTM PCR Amplification Kit in an autochonous sample from Spain. Forensic Sci. Int. 173, 241–245 (2007).
Kido, A., Dobashi, Y., Fujitani, N., Hara, M., Susukida, R., Kimura, H. et al. Population data on the AmpFlSTR identifiler loci in Africans and Europeans from South Africa. Forensic Sci. Int. 168, 232–235 (2007).
Maniatis, T., Fritsch, E. F. & Sambrook, J. Molecular Cloning. A Laboratory Manual, Cold Spring Harbor Laboratory Publications, New York, 458–459 (1982).
Walsh, P. S., Metzger, D. A. & Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based from forensic material. Biotechniques 10, 506–513 (1991).
Excoffier, L., Laval, G. & Schneider, S. Arlequin ver. 3.1: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 (2005).
Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139, 457–462 (1995).
Chakraborty, R., Fornage, M., Gueguen, R. & Boerwinkle, E. Population genetics of hypervariable loci: analysis of PCR based VNTR polymorphism within a population. in DNA Fingerprinting: Approaches and Applications (eds Burke, T., Dolf, G., Jeffreys, A.J. & Wolff, R.) 127–143 (Birkhauser, Verlag, Berlin, 1991).
Guo, S. W. & Thompson, E. A. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48, 361–372 (1992).
Nei, M., Tajima, F. & Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 19, 153–170 (1983).
Reynolds, J., Weir, B. S. & Cockerham, C. C. Estimation of the co-ancestry coefficient: basis for a short-term genetic distance. Genetics 105, 767–779 (1983).
Felsenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.6. (Department of Genome Sciences, University of Washington, Seattle, 2005).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 14, 406–425 (1987).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evo. 24, 1596–1599 (2007).
Shi, H., Dong, Y. L., Wen, B., Xiao, C. J., Underhill, P. A., Shen, P. D. et al. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am. J. Hum. Genet. 77, 408–419 (2005).
Wen, B., Xie, X., Gao, S., Li, H., Shi, H., Song, X. et al. Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am. J. Hum. Genet. 74, 856–865 (2004).
Aldenderfer, M. & Yinong, Z. The prehistory of the Tibetan plateau to the seventh century A.D.: perspectives and research from China and the West since 1950. J. World Prehist. 18, 1–55 (2004).
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. History and Geography of Human Genes.(Princeton University Press, Princeton, NJ, 1994).
Su, B., Xiao, J., Underhill, P., Deka, R., Zhang, W., Akey, J. et al. Y-chromosome evidencefor a northward migration of modern humans into eastern Asia during the last Ice Age. Am. J. Hum. Genet. 65, 1718–1724 (1999).
van Driem, G. Tibeto-Burman vs Indo-Chinese: implications for population geneticists, archeologists and prehistorians. in The Peopling of East Asia: Putting Together Archaeology, Linguistics and Genetics (eds Sagart, L., Blench, R. & Sanches-Mazas, A.) 81–106 (Routledge Curzon, London, 2005).
Su, B., Jin, L., Underhill, P., Martinson, J., Saha, N., McGarvey, S. T. et al. Polynesian origins: new insights from the Y-chromosome. Proc. Natl Acad. Sci. USA 97, 8225–8228 (2000a).
Jin, L. & Su, B. Natives or immigrants: modern human origin in East Asia. Nat. Rev. Genet. 1, 126–133 (2000).
Matisoff, J. A. Sino-Tibetan linguistics: present state and future prospects. Ann. Rev. Anthropol. 20, 469–504 (1991).
Thangaraj, K., Chaubey, G., Reddy, A. G., Singh, V. K. & Singh, L. Unique origin of Andaman Islanders: insight from autosomal loci. J. Hum. Genet. 51, 800–804 (2006).
Pérez-Miranda, A. M., Alfonso-Sánchez, M. A., Kalantar, A., García-Obregón, S., de Pancorbo, M. M., Peña, J. A. et al. Microsatellite data support subpopulation structuring among Basques. J. Hum. Genet. 50, 403–414 (2005).
Mateu, E., Rosa, M. A., Bernal, M., Zahra, B., Omar, A., Abdelaziz, S. et al. Genetic structure of north-west Africa revealed by STR analysis. Eur. J. Hum. Genet. 8, 360–366 (2000).
Takezaki, N. & Nei, M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144, 389–399 (1996).
Krithika, S., Maji, S. & Vasulu, T. S. A microsatellite guided insight into the genetic status of Adi, an isolated hunting-gathering tribe of Northeast India. PLoS ONE 3, e2549 (2008).
Nei, M. & Roychoudhury, A. K. Evolutionary relationships of human populations on a global scale. Mol. Biol. Evol. 10, 927–943 (1993).
Acknowledgements
We are grateful to the DNA donors and the investigators (Yu-Jiang Li, Jun-Feng Zhang) who helped in collecting the samples. We thank two anonymous reviewers for their valuable comments in earlier drafts of this paper. This study was supported by the National Natural Science Foundation (30760097), the Key Project of Chinese Ministry of Education (208138), the Postdoctoral Special Foundation of China (200902208), the Science Foundation of Ministry of Civil Affairs of the People's Republic of China (08xz02) and the Natural Science Foundation of Shanghai (10ZR1402200).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Kang, L., Li, S., Gupta, S. et al. Genetic structures of the Tibetans and the Deng people in the Himalayas viewed from autosomal STRs. J Hum Genet 55, 270–277 (2010). https://doi.org/10.1038/jhg.2010.21
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2010.21
Keywords
This article is cited by
-
Genetic structure of Tibetan populations in Gansu revealed by forensic STR loci
Scientific Reports (2017)
-
Detecting a hierarchical genetic population structure via Multi-InDel markers on the X chromosome
Scientific Reports (2016)
-
Genetic evidence for an East Asian origin of Chinese Muslim populations Dongxiang and Hui
Scientific Reports (2016)
-
History and possible mechanisms of prehistoric human migration to the Tibetan Plateau
Science China Earth Sciences (2016)