Abstract
Phylogenetic relationship and the population structure of 500 individuals from the Christian community of Lahore, Pakistan, were examined based on 15 autosomal short tandem repeats (STRs) using the AmpFℓSTR Identifiler Plus PCR Amplification Kit and our previously published Y-filer kit data (17 Y-STRs) of same samples. A total of 147 alleles were observed in 15 loci and allele 11 at the TPOX locus was the most frequent with frequency value (0.464). The data revealed that the Christian population has unique genetic characteristics with respect to a few unusual alleles and their frequencies relative to the other Pakistani population. Significant deviations from Hardy–Weinberg equilibrium were found at two loci (D13S317, D18S51) after Boneferroni’s correction (p ≤ 0.003). The combined power of discrimination, combined power of exclusion and cumulative probability of matching were 0.999999999999999978430815060354, 0.999995039393942 and 2.15692 × 10−17, respectively. On the bases of genetic distances, PCA, phylogenetic and structure analysis Lahore-Christians appeared genetically more associated to south Asian particularly Indian populations like Tamil, Karnataka, Kerala and Andhra Pradesh than rest of global populations.
Similar content being viewed by others
Introduction
Pakistan is a multiethnic country, harboring 217 million people, of whom the majority is Muslim according to the Pakistan Burea of Statistics1. Minority religious affiliates residing in Pakistan include Hindus, Christians, Ahmedis, Baha'is, Sikhs, Parsis, and Buddhists, amongst others. The Christian population comprises of 2.5 million (1.6%), making it the second largest religious minority of Pakistan2. Lahore, the capital of the Pakistani province of Punjab, is the second-most populous city in Pakistan (11.13 million) with a Muslim majority (97%) and a Christian minority (2%). Christianity was initially imported by Reverend Thomas Valpy who was appointed as the first Bishop of Lahore in 18773. Christians are considered to be descendants of a caste population of India4 and while they are thought to be a relatively closed population because of religious constraints, yet amiable relations are sustained with the majority population.
Short tandem repeats (STRs), also known as microsatellites, are repetitive sequences of DNA with a repeat motif of four to six base pairs and are almost universally employed as forensic identity markers because they are highly polymorphic and heterozygous, have short sequence lengths and are distributed throughout the human genome5,6 Although their mutation rates are significantly higher than those for single nucleotide polymorphisms (SNPs)7, they are none the less useful as genetic markers for population genetic studies, especially more recent genetic history8.
There have been many earlier studies of 15 autosomal STRs in various Pakistani populations except Christians. We emphasize that this population must be targeted as a whole, to understand the genetic context of Christians and its connection to the greater Eurasian continent. Hence, Lahore-Christian samples were evaluated based on fifteen autosomal STRs of Identifiler Plus Kit (Applied Biosystems) and already published data set of same male samples (YA004381)9 on 17 YSTRs (DYS438, DYS393, DYS385a⁄b, DYS389I⁄II, DYS458, DYS437, DYS391, DYS392, DYS635 (Y-GATA-C4), Y-GATA-H4, DYS19, DYS390, DYS439, DYS456, DYS448). To affirm phylogenetic affiliations of this population, data sets were compared with referenced populations as given in Table 1.
Materials and methods
Sample collection
About 3 mL blood was collected in EDTA vacutainer tubes from 500 unrelated Christian individuals residing Lahore, capital city of the Punjab province in Pakistan. Whatman blood stain cards were prepared for each sample with a unique sample ID that was henceforth used for processing.
DNA extraction and quantitation
Genomic DNA was isolated by an organic-extraction procedure described by Signer et al. (1988)10 and quantified on ABI7500 Real-Time PCR instrument (Applied Biosystems) using the Quantifiler Human DNA Quantification Kit (Applied Biosystems) following the recommended protocol11.
Amplification
DNA samples were diluted to the concentration of 1 ng/μL for PCR according to the recommended protocol for the AmpFℓSTR Identifiler Plus PCR Amplification Kit (Applied Biosystems)12. The DNA template (1 ng) was added to 2.4μL of Master Mix and 1.2μL Primer Set in a total reaction volume of 6 µL. PCR was performed in a GeneAmp9700 PCR System (Applied Biosystems). Thermal cycler conditions included an initial incubation for 11 min at 95 °C; 28 cycles of denaturation for 20 s at 94 °C, annealing/extension for 3 min at 59 °C and final extension for 10 min at 60 °C; and a final hold at 4 °C.
Genotyping
To perform genotyping on an ABI3730xl Genetic Analyzer (Applied Biosystems), 1µL of amplified product was added to 0.35µL GeneScan 500 LIZ size standard (Applied Biosystems) and 13µL highly deionized (Hi-Di) formamide. Data was analyzed using GeneMapper ID v3.2 to designate alleles in accordance with the Kit allelic ladder.
Quality control
The efficiency of the PCR amplification was monitored using Identifiler Plus Control DNA 9947A as a positive control and all reagents except DNA template as negative control. The STR analysis was conducted following the nomenclature recommendations of the DNA Commission of the International Society for Forensic Genetics (ISFG)13. The dataset was evaluated by the STRidER database13 with QC report reference number STR000284.
Population datasets used for comparison
The STR data of Lahore-Christians was compared with the available data of indigenous and global populations (supplementary Table 1) derived from published sources as summarized in Table 1.
Data analysis
Statistical parameters of forensic interest including power of discrimination (PD), matching probability (MP), observed (HO) and expected (HE) heterozygosities, polymorphism information content (PIC), typical paternity index (TPI), power of exclusion (PE) and allele frequencies were determined using modified Powerstats1.2 software30. A Hardy Weinberg equilibrium (HWE) test was performed using PowerMarker3.2531. The exact test for population differentiation was carried out by Arlequin3.5.2.2 software32. Phylogenetic and Principal Component analysis were executed using POPTREE33, MEGA-X34, Structure2.3.435 and PAST3.2636. Y-DNA haplogroups were also predicted by Whit Athey’s Haplogroup Predictor (https://www.hprg.com/hapest5/index.html)37 for the purpose of Y-lineage identification.
Ethical approval
All participants were introduced to this study and blood samples were collected with their Informed consent. The study was carried out in accordance with the relevant guidelines and regulations approved by the Ethical Committee of the Centre of Excellence in Molecular Biology, University of Punjab Lahore Pakistan (No. CEMB/AO/2289).
Results and discussion
Allelic frequencies and forensic parameters
A total of 147 alleles were observed over all loci and allele 11 at the TPOX locus was found to have the highest frequency of 0.46. Allelic frequencies at each locus are shown in Supplementary Table 2 while the parentage and forensic statistical parameters are in Supplementary Figure 1. Supplementary Table 3 shows five uncommon alleles (UCA) observed, together with most and least common alleles at each locus. Few alleles like 12.2, 14.2, 15.2, 16.2 at D19S433 and 9.1 at D7S820 were also reported to NIST STR Database. Polymorphism information content (PIC) was in the range of 0.623 (CSF1PO) to 0.841 (FGA) and the most discriminating marker was FGA with a PD value of 0.961. The observed heterozygosity varied from 0.656 (TPOX) to 0.868 (D8S1179) and the power of exclusion (PE) ranged from 0.364 (TPOX) to 0.731 (D8S1179). The power of discrimination (CPD), combined power of exclusion (CPE) and combined probability of matching (CPM) were 0.999999999999999978430815060354, 0.999995039393942 and 2.15692 × 10−17, respectively. Significant deviations from Hardy–Weinberg equilibrium (p < 0.05) were observed at two loci (D13S317, D18S51) after Boneferroni’s correction (p ≤ 0.003).
Interpopulation comparison
The allele frequencies at the 15 autosomal STRs in the Lahore-Christian population were compared with those from 16 other populations using population differentiation test as shown in Supplementary Table 4. Significant differences were observed after Boneferroni’s correction (p ≤ 0.0002) at 15/15 loci with Mongol, 14/15 African American26,27, Caucasian (12/15)27, Uganda Yousafzai and Kashmiri (11/15)16,18,28, Balochi, Punjabi, central India (7/15)14,23,38. While differences at small numbers of loci for Nepalese, Sindhi (3/15), Karnataka and Bangladeshi (2/15)21,24,25,39 were observed. However, there were no significant differences at any loci for the Tamil, Balmiki, and Kerala populations19,20,22.
Phylogenetic analysis
The neighbour-joining phylogenetic tree (Fig. 1A) illustrates genetic relationships between the Lahore-Christian population and 16 reference populations based on Fst corrected values of 15 autosomal STRs. Phylogenetic tree showed that Lahore-Christians appeared most closely associated to South Indians like Kerala and Tamil followed by Madhya Pradesh (Central Indian), Karnataka (Iyengar Brahmin) and Pakistani Punjabi. Other Pakistani Populations were distantly associated like Sindhi grouped with North Indian Balmiki; Balochi and Yousafzai Pathan shared genetic association to Caucasian, Uganda and African American. Similarly, a neighbour joining tree was constructed using our published 17-YSTRs data9 of studied population and 16 reference populations based upon RST p-values. Pairwise RST p-values were calculated through AMOVA using online YHRD tool. As illustrated in Fig. (1B) paternal lineage of Lahore-Christians shared branch with South Indian-Karnataka adjoining roots with Tamils, Andhra Pradesh and Bangladeshi followed by Punjabi. While Sindhi, Yousafzai and Balochi shared genetic association with Caucasians at the top. However, Madhya Pradesh (Central Indian) appeared distantly. Phylogenetic analysis shows Lahore-Christians population has a close genetic distance with the south Indian populations.
South India accounts for 21.47% of the community. It experienced a range of cross-cultural challenges between missionary Christianity and local converts40. Historically, the Tamil, Kerala and Karnataka populations belonged to the southern part of India and their culture is deeply rooted in Christians and Muslims41. Kerala is home to 22.07% of the total Christians in the country, followed by Tamil Nadu with 15.88%42. According to a 2011 census, Christians represent about 6% of the Tamil Nadu state population43 which also proclaim our phylogenetic analysis and migration history of Lahore-Christians. Moreover, it suggests that while South Indians and Pakistani Christians are geographically isolated, they have similar genetic origins.
Structure analysis
Although 15 autosomal STR markers have limited differentiation power to detect population structure but are efficient to some extent in differentiating Lahore-Christians from 9 other reference populations. Structure analysis was conducted employing Structure2.3.4 software using the admixture model with correlated allele frequencies without prior population information (USEPOPINFO = 0). Number of inferred clusters varied from 1 to 6 with three repetitions using 50,000 burnin and 100,000 MCMC simulation for each K. Results are intuitively depicted by bar plot as illustrated in Fig. 2A. All populations were partitioned into K colored segments depending on the value of K.
Whereas, K = 3 was the most suitable configuration based upon output posterior probability results inferred using the Structure Harvester44 as depicted in Fig. 2B. At K = 3 African American and Mongol were almost entirely filled with red and green component respectively. Lahore-Christians and Tamil shared blue color as major component structure in similar pattern that gradually diminished in next populations. Punjabi and Sindhi presented the mixture of green and blue components whereas Europeans (Caucasian, Romani) shared a mixture of red and green component to similar extent. While we may have expected Christians to exhibit some differentiation from the other Pakistani populations, it is not surprising that Lahore-Christians and South Asian Tamil are not differentiated by the STRs in the Identifiler panel using Structure.
Principal components analysis
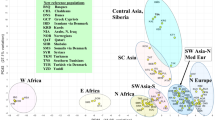
A PCA plot was constructed from autosomal STR allele frequencies (Supplementary Table 1) among Lahore-Christians, 4 indigenous reference populations (Fig. 3A) and global populations (Fig. 3B). In Fig. 3A Lahore-Christians signified as divergent population in lower right quadrant while rest of Pakistani populations clustered in upper right quadrant. Other global populations were scattered in the plot. In Fig. 3B Lahore-Christians were compared to Indian and 7 other world populations. It shows that studied population is relatively closer to South Indian populations (Karnataka, Kerala and Tamil) as compared to others. In Fig. 3A,B components 1 and 2 explain 55% and 46% of the variance respectively indicating genetic distances between populations.
(A) Principal component analysis (PCA) plot constructed from allele frequencies of 15 autosomal STR loci in the Lahore-Christian population, 4 indigenous populations (Punjabi, Sindhi, Yousafzai, Baloch) and 6 other populations. (B) PCA plot based upon allele frequencies of studied population, 5 Indian and 7 other reference populations. (C)The Multidimensional scaling (MDS) plot showed genetic relationships between Lahore-Christians, Pakistani, Indians and other populations.
Multidimensional scaling plot was generated based on haplotype data of YSTRs, to figure out Lahore-Christians paternal lineage (Fig. 3C). In this plot the Lahore-Christians remained tightly clustered with South Indian (Tamil, Karnatka, Andhra Pradesh), Central Indians (Madhya Pradesh) and Bangladeshi population. Balmiki was found distantly associated in the same quadrant whereas other populations including Pakistani, European American, Mongol, African American and Uganda scattered in different quadrants.
Y-DNA haplogroups
Haplotypes of Lahore Christians (n = 250) based on 17 –YSTRs using Whit–Athey’s algorithm were assigned to 7 haplogroups (L, Q, R, E1b1b, G2a, J2a1b, J2a1 x J2a1-bh). Other haplogroups like I1, G2c, I2a1, J2a1h I2b1, N and T were not observed in our samples. Whereas, L(40%), RIa(38%), E1b1b(25%), Q(23%) were found the most common haplogroups and accounted for most of its Y-lineage from south Asians. Our results also corroborated with the past reportings of the most frequent haplotypes from South Asia45.
The outcomes of phylogenetic analysis presented that Lahore Christians are most closely related to Indians particularly Tamil and might share common ancestors. Moreover, there are clear genetic variations between Christians and rest of the populations. It also supports historical records that, following the geographical migration from India to Pakistan, this population got eventually recognized as Christians46.
Lahore-Christians are primarily nomadic, poses conservative lifestyle, religious practices, extremely endogamous culture and traditional occupation as compared to other Pakistani population. Tracing their trail of migration and relatedness with world populations would provide a glimpse of primordial trajectory. Genetic affinity of Lahore-Christians to South Asian Indian populations and their common nomadic practices indicates historical genetic relatedness. Migratory events lead to subsequent separation of both populations. Relatively higher genetic distance to other Pakistani population were observed in our current study. Previous reports have also suggested genetic similarities of Tamils representing their common origin but minimal signature of gene exchange with other nomadic groups47.
However there were certain inconsistencies seen in side by side comparison at fewer population groups based on autosomal and YSTRs due to limited availability of their respective samples data. However, all these analyses clearly indicate that Lahore-Christian population has close genetic affiliation to South Indian population. Moreover, significant differences were observed between Lahore-Christians and other Pakistani populations except Punjabi that seems bit closer. This might also indicate that Lahore-Christians and Pakistani Punjabi diverged gradually from native South Indians following its geographical migration, which also corresponded with historical records48.
Conclusion
We have provided evidence that the Christian population in Lahore, Pakistan, forms a sub-population among Asian groups and has some unique genetic characteristics14,39,49. Results of inter-population differentiations, PCA, phylogenetic and structure analysis revealed that Lahore Christians have relatively close genetic relationships with south Asians particularly Indians. Being closely related to South Indians therefore it showed close resemblance to Tamil, Kerala, Andhra Pradesh and Karnataka populations. In this population, the 15 autosomal STRs and 17 YSTRs provide ample information for lineage characterization. This data would be useful for studies of genealogy, historical migration of Pakistani populations and database development. Genetic data obtained from autosomal and YSTR are in accord with human migration history of Indo-Pak populations. However, there is need of a detailed mitochondrial study to assign them mitochondrial haplogroups for maternal lineage identification.
References
Statistics, F. B. O. Government of Pakistan. Change (2007).
Victor, D. G., House, J. C. & Joy, S. A Madisonian approach to climate policy. Science 309, 1820–1821 (2005).
Hassan, F. Pakistan’s federal structure and the constitution of 1973. Muslim World 96, 269 (2006).
Pio, E. & Syed, J. in Faith-Based Violence and Deobandi Militancy in Pakistan 187–207 (Springer, 2016).
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435 (2004).
Willems, T. et al. The landscape of human STR variation. Genome Res. 24, 1894–1904 (2014).
Sun, J. X. et al. A direct characterization of human mutation based on microsatellites. Nat. Genet. 44, 1161 (2012).
Phillips, C. et al. Global patterns of STR sequence variation: sequencing the CEPH human genome diversity panel for 58 forensic STRs using the illumina ForenSeq DNA signature prep kit. Electrophoresis 39, 2708–2724 (2018).
Saleem, S. et al. Phylogenetic analysis and haplotype diversity in Christian residents of Lahore, Pakistan, using 17 Y-chromosomal STR loci. Int. J. Leg. Med. 11, 2017 (2019).
Signer, E., Kuenzle, C. C., Thomann, P. E. & Hübscher, U. DNA fingerprinting: improved DNA extraction from small blood samples. Nucleic Acids Res. 16, 7738 (1988).
Barbisin, M. et al. Developmental validation of the Quantifiler Duo DNA Quantification Kit for simultaneous quantification of total human and human male DNA and detection of PCR inhibitors in biological samples. J. Forensic Sci. 54, 305–319 (2009).
Applied Biosystems AmpFℓSTR Identifiler Plus PCR Amplification Kit—User Guide (2018). ThermoFisher Scientific, UK.
Bodner, M. et al. Recommendations of the DNA Commission of the International Society for Forensic Genetics (ISFG) on quality control of autosomal Short Tandem Repeat allele frequency databasing (STRidER). Forensic Sci. Int. Genet. 24, 97–102 (2016).
Shafique, M. et al. Genetic diversity of 15 autosomal STR loci in the population of Southern Punjab Pakistan. Forensic Sci. Int. Genet. 19, e1–e2 (2015).
Perveen, R. Forensic and Phylogenetic Characterization of Pakistani Population Using Uniparental and Biparental Genetic Markers (University of the Punjab, Lahore, 2018).
Mohapatra, B., KamalChauhan, U. & Thakur, B. Y. Anupuma Raina. Genetic analysis and evolutionary relationship of Jammu and Kashmir Muslim population with short tandem repeat loci. Int. J. Curr. Res. 8, 36398–36401 (2016).
Khan, A. A. et al. Genetic polymorphism of 15 autosomal short tandem repeats in Baloch population of Pakistan. Int. J. Leg. Med. 133, 775–776 (2019).
Batool, Z. et al. Genetic analysis of 15 autosomal STRs in Yousafzai population of Pakistan. Int. J. Leg. Med. 132, 1635–1636 (2018).
Balamurugan, K. et al. Genetic variation of 15 autosomal microsatellite loci in a Tamil population from Tamil Nadu, Southern India. Leg. Med. 12, 320–323 (2010).
Sreekumar, R. et al. Allelic frequency database of 15 polymorphic autosomal STRs in the Malayalam-speaking population of Kerala, India. Int. J. Leg. Med. https://doi.org/10.1007/s00414-020-02286-0 (2020).
Rajkumar, R. & Kashyap, V. Distribution of alleles of 15 STR loci of the Powerplex 16 multiplex system in four predominant population groups of South India. Forensic Sci. Int. 126, 173–177 (2002).
Ghosh, T. et al. Genetic diversity of autosomal STRs in eleven populations of India. Forensic Sci. Int. Genet. 5, 259–261 (2011).
Shrivastava, P., Jain, T. & Trivedi, V. B. Genetic polymorphism study at 15 autosomal locus in central Indian population. SpringerPlus 4, 566 (2015).
Ota, M. et al. Allele frequencies for 15 STR loci in Tibetan populations from Nepal. Forensic Sci. Int. 169, 234–238 (2007).
Hossain, T. et al. Genetic polymorphism studies on 22 autosomal STR loci of the PowerPlex Fusion System in Bangladeshi population. Leg. Med. 23, 44–46 (2016).
Zhan, X. et al. Forensic characterization of 15 autosomal STRs in four populations from Xinjiang, China, and genetic relationships with neighboring populations. Sci. Rep. 8, 1–7 (2018).
Hill, C. R., Duewer, D. L., Kline, M. C., Coble, M. D. & Butler, J. M. US population data for 29 autosomal STR loci. Forensic Sci. Int. Genet. 7, e82–e83 (2013).
Gomes, V. et al. Population data defined by 15 autosomal STR loci in Karamoja population (Uganda) using AmpF/STR Identifiler kit. Forensic Sci. Int. Genet. 3, e55–e58 (2009).
Anghel, A. et al. Genetic polymorphism data on 15 autosomal STR markers in a Western Romanian population sample. Leg. Med. 16, 238–240 (2014).
Tereba, A. Tools for analysis of population statistics. Profiles DNA 2, 14–16 (1999).
Liu, K. & Muse, S. V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129 (2005).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Takezaki, N., Nei, M. & Tamura, K. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol. Biol. Evol. 27, 747–752 (2009).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Pritchard, J. K., Wen, W. & Falush, D. Documentation for STRUCTURE software: Version 2. (2003).
Hammer, O., Harper, D. A. & Ryan, P. D. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9 (2001).
Athey, T. W. Haplogroup prediction from Y-STR values using a Bayesian-allele-frequency approach. J. Genet. Geneal 2, 34–39 (2006).
Khan, A. A. & Perveen, R. Nadeem Sheikh, Babar Hilal Ahmad Abbasi, Zunaira Batool, Muhammad Shahzad & Sana Kaleem.
Perveen, R., Shahid, A. A., Shafique, M., Shahzad, M. & Husnain, T. Genetic variations of 15 autosomal and 17 Y-STR markers in Sindhi population of Pakistan. Int. J. Leg. Med. 131, 1239–1240 (2017).
Doss, M. C. Indian Christians and the making of composite culture in South India. South Asia Res. 38, 247–267 (2018).
Bayly, S. Saints, Goddesses and Kings: Muslims and Christians in South Indian Society, 1700–1900 Vol. 43 (Cambridge University Press, Cambridge, 2004).
Reich, D., Thangaraj, K., Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009).
Collins, P. M. Christian Inculturation in India (Routledge, Abingdon, 2016).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Mahal, D. G. & Matsoukas, I. G. The geographic origins of ethnic groups in the Indian subcontinent: exploring ancient footprints with Y-DNA haplogroups. Front. Genet. 9, 4 (2018).
Pervaiz, H. & Mahmood, T. Mass conversion to Christianity: A case study of Chuhra Community in Sialkot Distric (1880–1930). Pak. Vis. 19, 40–59 (2018).
Watkins, W. et al. Genetic variation in South Indian castes: evidence from Y-chromosome, mitochondrial, and autosomal polymorphisms. BMC Genet. 9, 86 (2008).
Visaria, P. M. Migration between India and Pakistan, 1951–61. Demography 6, 323–334 (1969).
Ali, N., Coulson-Thomas, Y. M., Dixon, R. A. & Williams, D. R. Genetic variation comparison of 15 autosomal STR loci in an immigrant population living in the UK (British Pakistanis) with an ancestral origin population from Pakistan. Forensic Sci. Int. Genet. 9, e10–e12 (2014).
Acknowledgements
The authors gratefully acknowledge the volunteers who donated their blood, especially Muhammad Asim, the management of St. Oswald’s Church, St. Francis Xavier’s Catholic Church and St. Paul’s Anglican Catholic Church, Lahore Pakistan, for their cooperation and moral support.
Author information
Authors and Affiliations
Contributions
A.R. Did the research work. M.S. Performed Data evaluation and statistical analysis. A.A.S. Support for collecting the population samples and managerial issues. F.J. Manuscript write up and compilation. S.S. Did the sample collection and research work. F.Z. Prepared figures. D.M. Reviewed the manuscript critically regarding statistical analysis and English grammar. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rubab, A., Shafique, M., Javed, F. et al. Population genetic portrait of Pakistani Lahore-Christians based on 32 STR loci. Sci Rep 10, 18960 (2020). https://doi.org/10.1038/s41598-020-76016-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-76016-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.