Abstract
Transforming growth factor beta (TGFβ) signaling in breast cancer is selectively associated with pulmonary metastasis. However, the underlying mechanisms remain unclear. Here we show that Bcl-3, a member of the IκB family, serves as a critical regulator in TGFβ signaling to modulate breast cancer pulmonary metastasis. Bcl-3 expression was significantly associated with metastasis-free survival in breast cancer patients. Bcl-3 deletion inhibited the migration and invasion of breast cancer cells in vitro, as well as breast cancer lung metastasis in vivo. Bcl-3 was required for the expression of downstream TGFβ signaling genes that are involved in breast cancer lung metastasis. Bcl-3 knockdown enhanced the degradation of Smad3 but not Smad2 following TGFβ treatment. Bcl-3 could bind to Smad3 and prevent the ubiquitination and degradation of Smad3 protein. These results indicate that Bcl-3 serves as a promising target to prevent breast tumor lung metastasis.
Similar content being viewed by others
Main
Metastasis is the final and fatal stage of solid tumor progression and is responsible for most cancer-associated mortality. Transforming growth factor beta (TGFβ) signaling has been implicated in many steps of metastasis and is positively associated with the distant metastasis of various types of cancers, including breast cancer.1, 2, 3 Studies based on mouse models of breast cancer have shown that TGFβ signaling suppresses tumorigenesis but enhances the induction of epithelial–mesenchymal transition (EMT) and tumor invasion, consequently, promoting the seeding of lung metastases via genes like angiopoietin- like 4 (ANGPTL4).3, 4, 5 The activation of TGFβ signaling results in the phosphorylation of transcription factors Smad2 and Smad3, which accumulate in the nucleus in association with Smad4 and transactivate downstream target genes.6, 7 Recent reports have shown that Smad2 and Smad3 may have different roles in cancer metastasis. In particular, Smad3 enhances metastasis, whereas Smad2 suppresses metastasis.8, 9 Importantly, the reversible phosphorylation and ubiquitination of Smad2 and Smad3 proteins are indispensable processes that regulate TGFβ signaling.10, 11 Nedd4L has been reported to specifically recognize a TGFβ-induced phospho-Thr-ProTyr motif in the linker region of Smad2/3 via the WW domain, which results in Smad2/3 polyubiquitination and degradation.12 OTUB1 inhibits the ubiquitination of phospho-Smad2/3 by binding to and inhibiting the E2 ubiquitin-conjugating enzymes, independent of its deubiquitinase activity.13 The RBX1-SCFβ-TrCP E3 ligase complex targets activated Smad3 for nuclear export and ubiquitin-mediated degradation upon its association with the transcriptional co-activator p300.14 However, the proteins that participate in the regulation of Smad3 degradation are largely unknown.
B-cell lymphoma 3 (Bcl-3) is a proto-oncogene in the region adjacent to the t(14; 19) (q32; q13) translocation that was first identified in a subset of patients with chronic lymphocytic leukemia (CLL).15 Unlike other members of the IκB family, Bcl-3 cannot inhibit NF-κB nuclear translocation but binds to p50 and p52 dimers on DNA and provides the complex with transactivation ability.16, 17, 18 Bcl-3 has important roles in innate and adaptive immunity.19, 20 In addition, Bcl-3 expression has been shown to increase in a variety of hematopoietic and solid cancers, including breast cancer, to regulate tumor development and progression.21, 22, 23 The protein Cylindromatosis (CYLD) binds and deubiquitinates Bcl-3, thereby prevents its nuclear accumulation.26 In cylindromas and many other cancers with reduced CYLD expression,27 increased nuclear accumulation of Bcl-3 induces the transcription of target genes, such as cyclin d1 and N-cadherin, by interacting with NF-κB p50 and p52 homodimers.24, 25, 26, 27 These data suggest that Bcl-3 may serve as an oncogene by regulating the cell cycle or apoptosis at the gene and protein levels during cancer development. We have showed that Bcl-3 stabilizes c-Myc protein and promotes colorectal cancer development by regulating ERK signaling.22 Bcl-3 was recently reported to promote colorectal tumorigenesis through activation of AKT signaling.28 The above evidence reveals that Bcl-3 has many novel roles in tumor development and progression beyond being a trans-activator or trans-repressor. Recently, Bcl-3 has been demonstrated to regulate the metastasis of mouse breast cancer cells.21 Bcl-3 knockout MMTV-Neu mice resulted in reduced tumor lung metastasis with no effect on primary tumor growth.21 However, the underlying mechanisms of this metastasis remain unknown. Here we report that Bcl-3 functions as a critical regulator of TGFβ signaling by stabilizing Smad3 to promote the pulmonary metastasis of breast cancer.
Results
Bcl-3 expression is associated with the metastasis of breast cancer
We have previously reported that Bcl-3 was upregulated in human colorectal cancer compared with normal tissues.22 Bcl-3 expression has increased in breast cancers compared with normal mammary tissues.29, 30 Here we assessed Bcl-3 mRNA expression levels based on the Cancer Genome Atlas (TCGA) breast cancer (BRCA) data31 and the expression of Bcl-3 mRNA in tumors was much higher than tumor-matched normal tissues (n=99) (Figure 1a). Next, we detected the Bcl-3 expression of different breast cancer cell lines. Bcl-3 was highly expressed in malignant breast cancer cell lines but undetectable in MCF-10A, a normal mammary epithelial cell line (Figure 1b). Interestingly, the levels of Bcl-3 correlated with the pulmonary metastatic potential since highly metastatic cell lines LM2 expressed more Bcl-3 than cell lines with poor metastatic abilities like MCF-10A and MCF-7 (Figure 1b).These results suggest that Bcl-3 expression correlate with the pulmonary metastasis of breast cancer.
Bcl-3 expression is associated with the metastasis of breast cancer. (a) Expression of Bcl-3 in human breast cancer clinical specimens using the TCGA BRCA dataset. (b) Western blot analysis of Bcl-3 in a panel of breast cancer cell lines. (c) The metastasis-free survival curve for breast cancer patients with low (n=58, blue curve) versus high (n=58, red curve) Bcl-3 mRNA expression. (d) The metastasis-free survival curve for ER- breast cancer patients with low (n=41, blue curve) versus high (n=34, red curve) Bcl-3 mRNA expression. (e) The metastasis-free survival curve for ER- PR- breast cancer patients with low (n=31, blue curve) versus high (n=22, red curve) Bcl-3 mRNA expression. The patients were dichotomized on the basis of the mean value of Bcl-3 mRNA expression, and their survival curves were later analyzed
We then evaluated the elevated expression of Bcl-3 with metastatic progression and metastasis-free survival in breast cancer patients. Patients with breast cancers (n=116, P=0.025) that expressed higher mRNA levels of Bcl-3 had a significantly lower metastasis-free survival than patients whose tumors expressed lower levels of Bcl-3 (Figure 1c). The same results were found in estrogen receptor negative (ER−) (n=75, P=0.040, Figure 1d) and ER−PR− (progesterone receptors negative) (n=53, P=0.031, Figure 1e) breast cancers. These findings demonstrate a clinical significance of Bcl-3 in breast cancer and raise the need to further understand the function of Bcl-3 in breast tumor metastasis.
Loss of Bcl-3 inhibits the pulmonary metastasis of breast cancer in vivo
To investigate the effects of Bcl-3 on the pulmonary metastasis of breast cancer in vivo, we depleted Bcl-3 in different breast cancer cell lines using an inducible Tet-on lentivirus system.22 After infection with lentivirus, the cells were exposed to doxycycline (Dox) (1 μg/ml) for more than 4 days, and the expression of Bcl-3 was downregulated (Supplementary Figure 1c). Dox treatment did not affect the viability of cells (data not shown). We injected shRNA1-transduced LM2 cells, which were derived from parental MDA-MB-231 cells that exhibit highly pulmonary metastasis capability,22, 32 into nude mice via the tail vein, resulting in a substantial number of metastatic pulmonary nodules. The mice were fed with water containing Dox (2 μg/ml) to induce Bcl-3 silencing. The pulmonary metastatic ability was assessed based on the luciferase activity on days 0, 14, 28 and 42 (Figure 2a). Dox-inducible Bcl-3 knockdown cells exhibited lower photon flux than the control group. The mice were sacrificed after 7 weeks, and an analysis of the lung tissues indicated that Bcl-3 knockdown reduced the number of metastatic pulmonary nodules (Figure 2b).
Loss of Bcl-3 inhibits the pulmonary metastasis of breast cancer in vivo. (a) Bcl-3 knockdown LM2 luciferase cells were injected into female nude mice through the tail vein. One group was fed normal water, the other received 2 μg/ml Dox water. Representative lungs are shown by the BLI of the indicated cell lines at the indicated days after injection. Plots show a quantification of the luminescence signal as a function of time (n=6 for normal water; n=6 for Dox water). Values are mean±standard deviation. P<0.05 (two-tailed Student’s t-test). (b) Lung metastases were counted 8 to 9 weeks after the inoculation of the indicated cells through the tail vain. Representative H&E sections and photos are shown. (c) The lungs of FVB/N-Tg (MMTV-PyVT) 634Mul/J (ID:002374) and Bcl-3 knockout MMTV-PyVT female mice were harvested at necropsy and fixed with 4% polyoxymethylene; the number of lung metastasis foci were counted (n=8 for each group). Values are mean±S.D. P<0.05 (two-tailed Student’s t-test). Representative H&E sections and photos are shown. (d) WT bone marrow cells were transferred into lethally irradiated Bcl-3-sufficient or Bcl-3-deficient MMTV-PyMT mice. The lungs were harvested at necropsy after fixation with 4% polyoxymethylene, and the lung metastasis foci were counted (n=19 for Bcl-3-sufficient MMTV-PyMT mice; n=14 for Bcl-3-deficient MMTV-PyMT mice). Values are mean±S.D. P<0.01 (two-tailed Student’s t-test). (e) Cells from primary tumors of Bcl-3-sufficient or Bcl-3-deficient MMTV-PyMT mice were injected into the mammary pad of FVB mice. Lung metastases were counted 8 weeks after injection (n=9 for Bcl-3-sufficient MMTV-PyMT mice; n=11 for Bcl-3-deficient MMTV-PyMT mice). Values are mean±S.D. P<0.01 (two-tailed Student’s t-test). (f) The survival curve for mice in (e). *P<0.05 and **P<0.01 as determined by Student’s t-test
To confirm our observation in LM2 cells, we generated Bcl-3-sufficient and Bcl-3-deficient MMTV-PyMT mice, an aggressive transgenic mouse model of murine mammary adenocarcinoma development (MMTV-polyoma middle T (PyMT) mice).33 We found no gross histopathologic or quantitative differences between the groups based on primary tumor development or tumor burden (Supplementary Figure 2a). Strikingly, loss of Bcl-3 resulted in the attenuated pulmonary metastasis of breast cancer, as measured by significantly reduced numbers of metastatic foci and decreased tumor burden (Figure 2c). The pulmonary metastasis of mammary adenocarcinomas in MMTV-PyMT mice depends on hematopoietic cells, including CD4+ T cells, macrophages and TGFβ signaling.34 To verify the role of Bcl-3 in the pulmonary metastasis of cancer cells, we transferred wild-type (WT) bone marrow into lethally irradiated Bcl-3-sufficient or Bcl-3-deficient MMTV-PyMT mice. We observed increased primary tumor volumes but significantly reduced numbers of lung metastatic foci in Bcl-3-deficient MMTV-PyMT mice (Supplementary Figure 2b and Figure 2d). In addition, we injected cells from primary tumors of Bcl-3-sufficient and Bcl-3-deficient MMTV-PyMT mice into the mammary gland of FVB mice. Comparable volumes of primary tumors from both Bcl-3-sufficient and Bcl-3-deficient breast cancer cells were observed (data not shown). However, the number of metastatic pulmonary nodules was significantly reduced in mice injected with cancer cells from Bcl-3-null MMTV-PyMT mice compared with WT MMTV-PyMT mice (Figure 2e).
Importantly, the reduced lung tumor burden in Bcl-3-ablated lungs translated into significantly prolonged survival after tumor initiation (Figure 2f). These results demonstrate that Bcl-3 promotes pulmonary metastasis but not tumorigenesis in breast cancer.
To investigate the effects of Bcl-3 on pulmonary metastatic potential of breast cancer cells, we used in vitro systems to assess changes in cell motility and invasion. Knockdown of Bcl-3 with two shRNA sequences in both MDA-MB-231 and LM2 cells led to significantly reduced migration (Figures 3a and c,Supplementary Figure 6c and d) and Matrigel invasion (Figures 3b and d, Supplementary Figure 6c and d) ability. Wound-healing assay showed that Bcl-3 depletion significantly reduced cell migration compared with control cells in LM2 and 4T1 cells (Figures 3e and f and Supplementary Figure 6b). Together, these results suggest that Bcl-3 promotes the pulmonary metastasis of breast cancer cells by regulating the migration and invasion of breast cancer cells.
Loss of Bcl-3 inhibits the pulmonary metastasis of breast cancer cells. (a–d) Cell migration (a, c) and matrigel-transwell invasion (b, d) analysis of MDA-MB-231 cells (a, b) and LM2 cells (c, d), scale bar=50 μm. (e, f) Wound-healing assay of Bcl-3 knockdown LM2 cells (e) and 4T1 cells (f), scale bar=50 μm. Results are representative of three independent experiments. The data are expressed as mean±S.D. * represents P<0.05 and ** represents P<0.01 as determined by Student’s t-test
Bcl-3 regulates TGFβ target gene expression
To explore how Bcl-3 modulates the pulmonary metastasis of breast cancer, we detected a number of genes which are responsible for the enhanced pulmonary metastasis in LM2 cells, which has highly metastatic potential35 and autocrine production of TGFβ (Supplementary Figure 3b). Bcl-3 deletion significantly reduced the expression of TGFβ target genes ID1 (inhibitor of DNA binding 1), ID3 (inhibitor of DNA binding 1), MMP1 (matrix metallopeptidase 1), and COX2 (cytochrome c oxidase subunit II) in LM2 cells (Figure 4a). Next, globally profiled gene expression were performed to analyze genes affected by Bcl-3 depletion in MDA-MB-231 cells. Bcl-3 knockdown resulted in the differential expression of 1485 genes (>2-fold, P<0.05); 552 different genes were significantly upregulated (>2-fold increase, P<0.05), while 933 were significantly downregulated (<2-fold decrease, P<0.05) (Figure 4c). Gene ontology analysis showed that the most significantly reduced genes were associated with adherent junctions and the TGFβ signaling pathway, suggesting that Bcl-3 might be involved in TGFβ signaling (Figure 4b). To confirm that Bcl-3 was essential for TGFβ-related gene expression, we compared gene expression patterns in Bcl-3-sufficient and Bcl-3-deficient MDA-MB-231 cells after 24 h TGFβ stimulation. Bcl-3 knockdown caused a significant decrease in the expression of several TGFβ-responsive genes and pulmonary metastasis-related genes (Figure 4c). Microarray analysis revealed that 1049 genes were affected by TGFβ stimulation (428 induced and 621 repressed, >2-fold, Figure 4d), and knockdown of Bcl-3 after TGFβ stimulation affected 1458 genes (396 induced and 1062 repressed, >2-fold, Figure 4d). In addition, gene expression profiles revealed an overlap (355 genes) between affected genes of TGFβ stimulation and knockdown of Bcl-3 after TGFβ stimulation (Figure 4d). TGFβ primes LM2 cancer cells for pulmonary metastasis seeding via Angiopoietin-like potein 4 (ANGPTL4).3 The basal and TGFβ-induced ANGPTL4 mRNA levels were remarkably reduced in Bcl-3 depleted MDA-MB-231 and LM2 cells compared with control cells (Figure 4e). The functions of ID1 and ID3 are similar and redundant in regulating pulmonary metastasis,36 and Bcl-3 deletion reduced protein levels of both ID1 and ID3 (Figure 4f). Similarly, the TGFβ-induced levels of PAI-1, PTHrP, CTGF, and IL-11 (Supplementary Figure 3a) were dramatically decreased in Bcl-3-silenced MDA-MB-231 cells. ID1, ID3 and Snail, which are downstream targets of TGFβ signaling,37 have been reported to enhance EMT and tumor metastasis.36 We observed the protein levels of ID3 and Snail induced by TGFβ in Bcl-3-depleted MDA-MB-231 cells were profoundly reduced compared with the control cells. TGFβ did not induce ID1 expression in MDA-MB-231 cells, TGFβ instead reduced it when Bcl-3 was absent (Figure 4f). Consistently, Bcl-3 knockdown with shRNA1 sequence led to a significant reduction in wound healing induced by TGFβ in MDA-MB-231 cells (Figure 4g). In LM2 cells not MDA-MB-231 cells, Bcl-3 knockdown led to the reduction in wound healing even without TGFβ treatment (Figure 3e), the reason may be the higher autocrine production of TGFβ in LM2 cells (Supplementary Figure 3b). Taken together, these results indicate that Bcl-3 promotes breast cancer metastasis and invasion by regulating TGFβ signaling.
Bcl-3 regulates TGFβ target gene expression. (a) qRT-PCR analysis of metastasis-related genes in Bcl-3 knockdown LM2 cells. (b) Gene ontology analysis shows altered signaling pathway. (c) Heat map of the altered genes shared between the four profiles; red shows induced and green shows repressed genes, log2-based scale. MDA-MB-231 cell RNA samples were collected after TGFβ stimulation for 24 h. (d) Venn diagram representation of differentially expressed genes in the two groups: genes affected by TGFβ stimulation and genes affected by Bcl-3 depletion after TGFβ stimulation. (e) qRT-PCR analysis for ANGPTL4 in the indicated MDA-MB-231 and LM2 cells with or without TGFβ stimulation for 24 h. The results are the means±S.E.M.s for each cohort (n=3). Student’s t-test was used. P<0.05. (f) Immunoblots for Bcl-3, Snail, ID1 and ID3 in the indicated MDA-MB-231 cells under TGFβ stimulation. (g) Wound-healing assay carried out on the indicated MDA-MB-231 cells under TGFβ stimulation, scale bar=50 μm
Bcl-3 regulates TGFβ signaling by stabilizing Smad3 protein
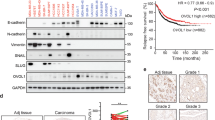
To characterize how Bcl-3 regulates TGFβ signaling, we examined the phosphorylation of Smad proteins in control and Bcl-3-silenced MDA-MB-231 cells. Bcl-3 knockdown did not influence the early phosphorylation and total levels of Smad2 or Smad4 induced by TGFβ. Surprisingly, the phosphorylation and total protein levels of Smad3 decreased after TGFβ treatment in Bcl-3 knockdown cells, and these changes were similar with ID1 protein expression (Figure 5a,Supplementary Figures 3c, d). Consistent results were obtained in Bcl-3-depleted LM2, MCF-7 cells and the mouse breast cancer cell line 4T1 (Supplementary Figures 3e-g). The results above indicate the correlation between Bcl-3 and Smad3. Then we considered the protein level of Bcl-3 and Smad3 in some breast cancer cells with different metastasis potential. Our results showed that these two proteins expressed consistently in these cells, as MCF-10A exhibited lowest Bcl-3 and Smad3 while LM2 cells showed the highest levels of Bcl-3 and Smad3 (Figure 5b). To explore whether Bcl-3 modulates Smad3-mediated gene transcription, we utilized a TGFβ reporter plasmid38 to measure the TGFβ-induced transcriptional activity. Knockdown of Bcl-3 with two shRNA sequences significantly inhibited the TGFβ reporter luciferase activity induced by TGFβ in MDA-MB-231 (Figure 5c) and MCF-7 (Supplementary Figure 3h) cells. These data indicate that Bcl-3 regulates TGFβ signaling by affecting the protein levels of Smad3 but not Smad2 or Smad4.
Bcl-3 regulates TGFβ signaling by stabilizing Smad3 protein. (a) Immunoblots for Smad2, phospho-Smad2, Smad3 and phos-Smad3 in MDA-MB-231 cells under TGFβ stimulation. (b) Immunoblots for Bcl-3 and Smad3 in MCF-10A, MCF-7, MDA-MB-231 and LM2 cells. (c) The knockdown of Bcl-3 inhibited the TGFβ reporter luciferase activity in MDA-MB-231 cells. MDA-MB-231 cells with two different shRNA sequences were co-transfected with a TGFβ reporter luciferase plasmid and a Renilla luciferase normalization control. After stimulation with TGFβ, the cell lysates were collected, and the firefly luciferase and Renilla activities were measured with a dual-luciferase reporter system. The results are the means±S.E.M.s for each cohort. (n=3). *P<0.05. (d) qRT-PCR analysis of Smad3 in Bcl-3 knockdown MDA-MB-231 cells treated with TGFβ. (e) Immunoblots for Smad3 in Bcl-3 knockdown MDA-MB-231 cells treated with MG132 for 6 h and TGFβ. (f) WT ubiquitination, K63-linked ubiquitination and K48-linked ubiquitination of Smad3 in MDA-MB-231 cells after MG132 and TGFβ stimulation. The immunoblot analysis of immunoprecipitated Smad3 for the presence of ubiquitin, K48-linked ubiquitin and K63-linked ubiquitin, and Smad3 is shown. (g) Immunoblots for Smad3, Bcl-3 and GAPDH in RBX1 (up) or β-TrCP (bottom) knockdown MDA-MB-231 cells treated with TGFβ. (h) Cell migration ability assessed by a transwell invasion analysis (left) and migration assay (right) in Smad3 overexpressing, Bcl-3-silenced MDA-MB-231 cells
The depletion of Bcl-3 in breast cancer cells reduced the steady-state levels of Smad3 protein upon TGFβ stimulation but no change in the Smad3 mRNA level (Figure 5d). These data suggest that Bcl-3 regulates the Smad3 levels via a posttranscriptional mechanism. Furthermore, the treatment of cells with the proteasome inhibitor MG132 restored the Smad3 protein levels decreased by Bcl-3 knockdown after TGFβ treatment, confirming that Smad3 was degraded by the ubiquitin–proteasome system (Figure 5e). These data indicate that Bcl-3 depletion increases the proteasome-dependent degradation of Smad3 following TGFβ stimulation. Indeed, Smad3 was found to be ubiquitinated in a K48-linked but not K63-linked manner in Bcl-3-depleted MDA-MB-231 cells after MG132 treatment and TGFβ stimulation (Figure 5f and Supplementary Figure 3j).
An E3 ubiquitin ligase complex RBX1-SCFβ-TrCP is responsible for the ubiquitination of TGFβ-activated Smad3.14 This E3 ligase complex is composed of RBX1 (RING box protein 1), Skp1, Cullin1 and β-TrCP.14 However, the depletion of RBX1 or β-TrCP could not lead to an accumulation of Smad3 in Bcl-3-sufficient or Bcl-3-silenced MDA-MB-231 cells following TGFβ stimulation (Figure 5g). These results indicate that Bcl-3 stabilizes Smad3 via the ubiquitination dependent degradation pathway, but not regulated by the E3 ligase complex RBX1-SCFβ-TrCP.
In addition, the significantly reduced invasion and wound-healing migration rates caused by the knockdown of Bcl-3 were partially rescued by Smad3 overexpression in Bcl-3-depleted LM2 cells (Figure 5h). Rescue experiments in vitro further confirm that Bcl-3-depletion induced decreases of Smad3 protein in breast cancer cells contribute to reduced migration and invasion ability in vitro and lung metastasis in vivo.
Bcl-3 stabilizes Smad3 by forming a complex with Smad3
Phosphorylation at different sites of Smad3 contributes to its stability.39 To determine if Bcl-3 stabilized the Smad3 protein level via influencing Smad3 phosphorylation, we measured the phosphorylation levels at several Smad3 phosphorylation sites after TGFβ stimulation in LM2 and MDA-MB-231 cells, including phosphor-Smad2/Smad3 Thr8, phosphor-Smad2/Smad3 Thr179, phosphor-Smad3 Ser204, phosphor-Smad3 Ser208, phosphor-Smad3 Ser213 and phosphor-Smad3 Ser423/425. The phosphorylation at Ser204, which is located in the linker region of Smad3, was significantly reduced in Bcl-3-depleted LM2 cells (Supplementary Figure 4a), but not in MDA-MB-231 (Supplementary Figure 4b). Moreover, we transfected Bcl-3-silenced MDA-MB-231 cells with WT Smad3, a Smad3 MH2 domain phosphorylation mutant called dSSVS, or a Smad3 linker region phosphorylation mutant called Smad3 EPSM. Similar to WT Smad3, decreased levels of mutated Smad3 proteins were detected in Bcl-3-depleted cells upon TGFβ treatment (Supplementary Figure 4c). Taken together, these results indicate that the phosphorylation of Smad3 is not required for its regulation by Bcl-3 in TGFβ signaling, even though Bcl-3 affected the Smad3 phosphorylation at Ser204 in LM2 cells.
Given that Bcl-3 was selectively needed to stabilize Smad3 not Smad2, we hypothesized that Bcl-3 forms a complex with Smad3 to prevent the degradation of Smad3. First, we co-transfected expression vectors encoding Bcl-3 and Smad3 into 293 T cells and immunoprecipitated the lysates with antibodies directed against either protein tag. Immunoblots revealed that Bcl-3 was present in Smad3 immunoprecipitates and vice versa (Figures 6a and b). Further mapping revealed that an N-terminal deletion mutant of Bcl-3 (deN) could not interact with Smad3 (Figure 6d), while the C-terminal (MH2 domain) deletion (deC) mutant of Smad3 could not bind with Bcl-3 (Figure 6e). Consistently, Bcl-3 did not interact with endogenous Smad2 (Figure 6c). These results demonstrate that Bcl-3 binds to the MH2 domain of Smad3 to stabilize Smad3 by disrupting the degradation of Smad3.
Bcl-3 stabilizes Smad3 by forming a complex with Smad3. (a) Co-IP of Flag-tagged Smad3 (Smad3-Flag) and hemagglutinin-tagged-Bcl-3 (Bcl-3-HA) expressed in HEK293T cells. (b) Co-IP of Flag-Myc-tagged Bcl-3 (Bcl-3-Flag-Myc) and Flag-tagged Smad3 (Smad3-Flag) expressed in HEK293T cells, cell lysates were immunoprecipitated with Myc antibodies. (c) HA-Bcl-3 was expressed in HEK293T cells. Forty-eight hours after transfection, the cells were lysed, and lysates were immunoprecipitated with HA antibodies. (d) Co-IP of Smad3-HA and Bcl-3 truncated expression vectors (WT: 1–446 amino acids; deN30: 31–446 amino acids; deN:125–446 amino acids; deC:1–330 amino acids) in HEK293T cells and cell lysates were immunoprecipitated with Flag (upper) and HA (bottom) antibodies. (e) Co-IP of Bcl-3-Flag and Smad3 truncated expression vectors (WT: 1–426 amino acids; deN: 138–426 amino acids; deC: 1–233 amino acids) in HEK293T cells and cell lysates with Flag antibodies. * represents nonspecific band. The deN mutant has the same molecular weight with nonspecific band
Without TGFβ treatment, Smad3 was located in the cytoplasm, whereas Bcl-3 were predominantly detected in the nucleus. Under TGFβ stimulation, Smad3 trans-located into the nucleus where Bcl-3 co-localized with Smad3. When Bcl-3 was knocked down, the levels of Smad3 in the nucleus profoundly decreased after TGFβ treatment. An immunofluorescence analysis revealed that Bcl-3 interacted with Smad3 in the nucleus, protecting Smad3 from degradation (Supplementary Figure 5).
Discussion
TGFβ signaling has a dual role in breast cancer tumorigenesis:40, 41 it inhibits tumor proliferation in the early stage, and promotes tumor metastasis via EMT in the advanced stages of carcinogenesis. Our study showed that Bcl-3 regulated the process of breast cancer pulmonary metastasis without affecting tumor proliferation. We found the elevated expression of Bcl-3 correlated with metastatic progression and metastasis-free survival in breast cancer patients. Moreover, Bcl-3-deficient MMTV-PyMT mice showed a significantly reduced number of pulmonary metastatic foci. Bcl-3 knockdown dramatically reduced the migration and invasion of breast cancer cells.
Bcl-3 was recently shown to be able to promote the metastasis of ErbB2-positive mammary tumors,21 but little is known about the mechanism. In our study, we found that Bcl-3 regulated breast cancer pulmonary metastasis in breast cancer cells by modulating TGFβ signaling via the stabilization of Smad3 and other metastasis-related proteins such as ID1 (see discussion below).
Researchers have investigated the specific functions of Smad2 and Smad3 in TGFβ-induced signaling in breast cancer cells in vitro and in a mouse model of breast cancer metastasis. The knockdown of Smad3 in MDA-MB-231 cells resulted in the prolonged latency and delayed growth of metastases, whereas Smad2 knockdown resulted in a more aggressive phenotype compared with control cells.9 Therefore, we demonstrated that Bcl-3 serves as a critical regulator in stabilizing Smad3, but not Smad2 or Smad4, upon TGFβ stimulation. This function is crucial for the TGFβ-dependent cell migration, invasion and pulmonary metastasis of breast cancer.
Notably, Smad3 overexpression only partially rescued the deficiency caused by Bcl-3 loss, suggesting that other mechanisms might be involved in the regulation of the pulmonary metastasis of breast cancer by Bcl-3. First, Bcl-3 might regulate EMT independently of Smad3. Indeed, we found that Bcl-3 depletion in some breast cancer cell lines, such as MDA-MD-231 and 4T1 cells, led to mesenchymal-to-epithelial transition (MET) induction, there was as demonstrated by reduced F-actin polarity (Supplementary Figure 4d) and an obvious morph-change from fibroblast-like to round-like (Supplementary Figure 4e). Second, Bcl-3 may regulate the stability of other metastasis-related proteins in addition to Smad3. For example, we observed that the protein levels of ID1 were regulated by Bcl-3, especially upon TGFβ treatment. However, we did not detect the interaction of Bcl-3 with these proteins. The mechanisms underlying the Bcl-3 regulation of metastasis need to be further investigated.
TGFβ signaling is mainly determined by the expression level and activity of the effector proteins Smad2 and Smad3.39 After TGFβ stimulation, Smad3 can be recognized by the E3 ligase RBX1 at its MH2 domain and then degraded by the ubiquitin–proteasome pathway.14 Smad3 is phosphorylated at Ser208 by ERK MAP kinase upon EGF treatment, and ERK phosphorylation inhibits Smad3 activity because the mutation of the ERK phosphorylation sites increases the ability of Smad3 to stimulate a Smad target gene.42 We found that the phosphorylation at Ser204, located in the link region of Smad3, was significantly reduced in Bcl-3-depleted cells, which might be accounted for by our previous finding that Bcl-3 depletion reduced ERK activation22 (data not shown). However, the Smad3 linker region phosphorylation did not differ between mutant EPSM and control cells after TGFβ treatment. These results indicate that the stabilization of Smad3 by Bcl-3 does not depend on the phosphorylation of the Smad3 linker domain.
Importantly, Bcl-3 directly interacts with the MH2 domain of Smad3, which is consistent with reports that DEAR1 and AKT bind to the MH2 domain of Smad3 but do not interact with Smad2.43, 44, 45 MH2 domain of Smad3 is the binding site of E3 ligase or adapter such as DEAR1 that promote Smad3 ubiquitination and degradation. The binding of Bcl-3 with MH2 domain of Smad3 but not Smad2, suggest that Bcl-3 stabilizes Smad3 through competitively preventing the interaction of Smad3 with its E3 ligase, probably other proteins such as DEAR1. Therefore, we propose that Bcl-3 stabilizes Smad3, but not Smad2 or Smad4, by disrupting the interaction of Smad3 and its E3 ligase, preventing the degradation of Smad3 by the ubiquitin–proteasome pathway. However, the depletion of RBX1 or β-TrCP, the components of the E3 ubiquitin ligase complex which is responsible for the ubiquitination of TGFβ-activated Smad3, could not lead to an accumulation of Smad3 in MDA-MB-231 cells whether TGFβ stimulation or not, maybe other E3 ubiquitin ligase participate in this process. This process is not associated with the phosphorylation of Smad3, but depend on the binding affinity of Bcl-3 and Smad3.
In summary, our study uncovered a novel mechanism by which Bcl-3 stabilizes Smad3 protein during TGFβ signaling. Because TGFβ signaling plays important roles in breast cancer pulmonary metastasis,46 our findings provide a promising new drug target for the prevention and therapy of breast cancer pulmonary metastases.
Materials and Methods
Cell lines
The human breast cancer cell lines HEK293T, MDA-MB-231, MCF-10A, MCF-7, were purchased from ATCC (Rockville, MD, USA). LM2 was a kind gift from Dr Joan Massagué. The mouse breast cancer cell line 4T1 was purchased from SGST (Shanghai, China). HEK293T, MDA-MB-231, LM2 and MCF-7 were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, HyClone, Logan, UT, USA) supplemented with 10% FBS, 1% penicillin and streptomycin in a 5% CO2 humidified atmosphere at 37 °C. 4T1 cells were maintained in RMPI-1640 medium (HyClone, Logan, UT, USA) supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin.
Patient data
Breast cancer tissues were collected from patients at Qilu Hospital of Shandong University. All of these tumors were histologically confirmed as breast carcinomas. Informed consent for participation in this study was obtained from all patients before their surgeries. And human subject studies were approved by the institutional review boards of Qilu Hospital with informed patient consent.
Lentivirus-delivered shRNA gene knockdown and overexpression
To knockdown the Bcl-3 gene in breast carcinoma cell lines, two different Bcl-3 shRNA sequences were cloned into the pTRIPZ plasmid (Open Biosystems, RHS4750, Huntsville, Alabama, USA), a tet-on lentiviral vector, according to the manufacturer’s instructions (V3THS_407972), respectively. The third shRNA sequence targeting Bcl-3 was cloned into plvx-shRNA plasmid. The non-silencing lentiviral shRNA vector was used as a control. The lentivirus were packaged using psPAX2 and pMD2G, a three-plasmid system. To obtain stable cell lines, lentivirus supernatant was added to MDA-MB-231, LM2 and 4T1 cells, followed by screening with 1 μg/ml puromycin for MDA-MB-231, 0.5 μg/ml puromycin for MCF-7, and 14 μg/ml puromycin for 4T1 cells for 2 weeks. The expression of Bcl-3 was downregulated in these cell lines when treated for longer than 4 days with 1 μg/ml doxycycline (Dox), an analog of tetracycline, in the culture medium. To overexpress Smad3 in Bcl-3 knockdown cells, Smad3 was cloned into the pLVX-IRES-ZsGeen1 plasmid. Lentivirus supernatant was added to the culture medium of Bcl-3 knockdown cells. We assessed the infection rate using a fluorescence microscope.
TCGA data analysis
The RNA-seq dataset were retrieved from the Cancer Genome Atlas (TCGA) database and only appropriate tumor-normal matched samples were included (sample size=99). RSEM normalized results from TCGA were applied to downstream analysis, and we manually checked expression level of Bcl-3 in these samples.
Plasmids and antibodies
Flag-tagged Bcl-3, or HA-tagged Smad3 were cloned into pcDNA3.1 and pcDNA3.0. All PCR products were confirmed by sequencing. The following antibodies were used: anti-Bcl-3, c-Myc, cyclinD1, p27, p21, vimentin, E-cadherin, N-cadherin, ID1, FLAG, HA, ubiquitin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), anti-Snail, ID3, RBX1 (Proteintech), anti-phos-ERK, phos-AKT, AKT, phos-Smad3, Smad3, phos-Smad2, Smad2, Lamin A/C (Cell Signaling Technology, Boston, MA, USA), anti-GAPDH, donkey anti-Goat IgG (HRP), goat anti-Mouse IgG (HRP), goat anti-Rabbit IgG (HRP) (KANGCHEN, Shanghai, China), anti-Actin (Sigma-Aldrich, St. Louis, MO, USA), anti-K63-ubiquitin, K48-ubiquitin (Millipore, Billerica, MA, USA), donkey anti-goat coupled to AlexaFluor®488, and donkey anti-mouse or rabbit IgG coupled to AlexaFluor680 (Invitrogen, Carlsbad, CA, USA). Anti-Smad3 Ser423/425 (Cell Signaling Technology), anti-Smad2/3 pT8, anti-Smad2/3 pT179, anti-Smad3 pS204, anti-Smad3 pS208, anti-Smad3 pS213 were kind gifts from Dr Liu Fang.
Immunoprecipitation and immunoblotting
Confluent cells in 10-cm tissue culture dishes were washed with PBS containing 10 mM CaCl2 and then lysed in 1 ml of lysis buffer (20 mM Hepes (pH 7.4), 12.5 mM β-glycerophosphate, 0.5% Triton-X-100, 150 mM NaCl, 1.5 mM MgCl, 2 mM EGTA, supplemented with phosphatase and protease inhibitors) and incubated on ice for 30 min before clearing by centrifugation at 14 000 r.p.m. for 15 min. The proteins from cell extracts were immunoprecipitated out using protein A-Sepharose (GE, Fairfield, CT, USA) coated with specific antibodies. Following overnight incubation at 4 °C, immunocomplexes were collected and washed four times with lysis buffer. Bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE. Immunoblotting was carried out using standard procedures, and immune-reactive proteins were visualized by enhanced chemiluminescence (ECL). For ubiquitination assay, MG-132 (Beyotime, Shanghai, China s1748, 100 mmol/l for 12 h) were used to inhibit proteasome activity.
Mice
All animals were housed and maintained in pathogen-free conditions. All animal experiments were performed in compliance with the guide for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of the Shanghai Institutes for Biological Sciences at the Chinese Academy of Sciences. Female BABL/c nude mice were purchased from the Shanghai Laboratory Animal Center at the Chinese Academy of Sciences in Shanghai. The Bcl-3 knockout (KO) mice have been described previously.47 Female Bcl-3−/− mice were crossed with male FVB mice that were transgenic (+/−) for the PyMT antigen under the control of the MMTV promoter. Genotyping for the PyMT transgene was performed by PCR. Female mice from this cross that were PyMT+/− were saved for further analysis. The mice were sacrificed at 15 weeks of age, and the whole mammary glands, tumors and/or lungs were excised.
Histology and immunohistochemistry
Anti-Bcl-3 antibodies (Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA) were used as the primary antibodies. IHC analyses of diaminobenzidine staining were performed using an HRP kit (UltraTek; Scytek, Logan, UT, USA). The diaminobenzidine-stained specimens were visualized using a general optical microscope with a camera (Carl Zeiss, Oberkochen, Germany). Hematoxylin and eosin (H&E) staining of the tissue was conducted using a MIRAX scan (Carl Zeiss). Images were processed with equivalent parameters using ZEN Light Edition software (Carl Zeiss).
Immunofluorescence
Immunofluorescence was analyzed as described previously.48 The cells were plated at 3 × 105 cells per 35 mm dish onto coverslips and washed 3 times in 100 mM PBS before being fixed in cold acetone (5 min, room temperature). The cells were blocked with PBS containing 1% FBS and 0.3% Triton-X-100 for 30 min. The proteins were localized using the primary antibodies overnight at 4 °C. The cells were washed and incubated with secondary antibodies coupled to AlexaFluor®488 or AlexaFluor®680 (1:500, 2 h, room temperature). The cells were then observed with a Zeiss laser-scanning confocal microscope (LSM Meta 510) using a Fluar Plan Apochromat × 63 oil immersion objective (numerical aperture 1.4) or a Plan Apochromat × 100 oil immersion objective (numerical aperture 1.4). The images were collected at a zoom of 2–3 and an iris of <3 μm. Single sections are shown. The images were processed (colored and merged) using the Zeiss (LSM 510) software. More than 20 cells were analyzed for each experiment.
Luciferase assay
TGFβ reporter luciferase plasmid38 was provided by Dr Ying E Zhang of NCI. MDA-MB-231 cells were cultured in 24-well dishes and transfected with 50 ng of TGFβ reporter luciferase plasmids and 20 ng of Renilla plasmids using Lipofectamine 2000. After 24 h of transfection, the cells were lysed in reporter lysis buffer (Promega, Madison, WI, USA). The cell extracts were collected, and the firefly luciferase and Renilla activities were measured with a Dual-Luciferase Reporter System (Promega).
RNA extraction and real-time PCR
RNA was isolated from cell lines or patient samples using TRIzol Reagent (Invitrogen, 15596-018) according to the manufacturer’s protocol. To obtain cDNA, reverse transcription was performed using Transcript First Strand Synthesis Supermix (TransGen Biotech, Beijing, China AT301) according to the manufacturer’s instructions, using 1 μg of RNA as a template. All qRT-PCRs were performed using a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA), and all qRT-PCR reagents and consumables were purchased from Applied Biosystems. For each reaction, 1 μl of RT product was added to 10 μl of 2X SYBR Green Gene Expression PCR Master Mix and 1 μl of pre-designed and synthesized forward and reverse primer/probe mix. Each sample was analyzed in triplicate. Relative quantification (RQ) was derived from the difference in the cycle threshold (Ct) between the target gene and GAPDH (ΔCt) compared with control cell lines using the equation RQ=2−ΔΔCt. Error bars represent the standard deviation (SD), and the significance of differences was calculated using a one-tailed, unpaired t-test. The sequences of the primers are listed in Supplementary Table 1. The patients were dichotomized on the basis of the mean value of Bcl-3 mRNA expression, and their survival curves were later analyzed.
Wound-healing assay
Cell monolayer in 6-well plates was artificially scratched with 10 μl pipette tips. The wound areas were photographed 0 and 20 h after scratching and measured using a caliper. The wound-closure percentages were calculated using the following formula: (1-(current wound size/initial wound size)) × 100.
In vitro invasion assays
In vitro invasion assays were conducted using Transwell inserts (Costar, Cambridge, MA, USA) containing 8-μm pore-size polycarbonate membrane filters in 24-well culture plates. The upper surface of the filter was coated with 20 μg of Matrigel (Becton Dickinson, Bedford, MA, USA) per filter. The Matrigel was dried and reconstituted at 37 °C into a solid gel on the filter surface. After starving in FBS-free DMEM overnight, 1 × 105 MDA-MB-231 cells were seeded in the upper chamber in 200 μl of 0.1% BSA DMEM. The lower chamber was flooded with 500 μl of 10% FBS DMEM. After 18 h, the wells were fixed with 4% polyoxymethylene and stained with crystal violet. Cells that invaded the lower surface of the filter were counted in 6 random fields under a light microscope at high magnification. Experiments were repeated at least in triplicate.
In vivo metastasis assays
For experimental metastasis assays, age-matched female nude mice were injected with 2 × 105 LM2 cells stably expressing an inducible Bcl-3 knockdown vector in PBS via the tail vein. After 7 weeks, the mice were sacrificed, and the lungs were collected and immobilized in Bouin’s solution to count the number of foci and stain the tissues with hematoxylin and eosin (H&E). To observe living bodies, 2 × 105 LM2 with the indicated constructs were injected into age-matched female nude mice via the tail vein, and D-luciferin was retro-orbitally injected at days 1, 14, 28 and 42. The photon flux (photons per second) was recorded.
Gene expression profiling
The total RNA was extracted using TRIzol Reagent (Cat No.15596-018, Life technologies, Carlsbad, CA, USA) following the manufacturer’s instructions and checked for a RIN number to inspect RNA integrity with an Agil manufacturer’s ent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, USA). Qualified total RNA was further purified with the RNeasy micro kit (Cat No. 74004, QIAGEN, GmBH, Germany) and RNase-Free DNase Set (Cat No. 79254, QIAGEN, GmBH, Germany). The total RNA was amplified, labeled and purified using a GeneChip 3’IVT Express Kit (Cat No. 901229, Affymetrix, Santa Clara, CA, USA) following the manufacturer’s instructions to obtain biotin-labeled cRNA. Array hybridization and washing was performed using a GeneChip® Hybridization, Wash and Stain Kit (Cat No. 900720, Affymetrix, Santa Clara, CA, USA) in a hybridization oven 645 (Cat No. 00-0331-220 V, Affymetrix, Santa Clara, CA, USA) and a fluidics station 450 (Cat No. 00-0079, Affymetrix, Santa Clara, CA, USA) following the manufacturer’s instructions. The slides were scanned by a GeneChip Scanner 3000 (Cat No. 00-00212, Affymetrix, Santa Clara, CA, USA) using the Command Console Software 3.1 (Affymetrix, Santa Clara, CA, USA) and default settings. The raw data were normalized using the MAS 5.0 algorithm, Gene Spring Software 11.0 (Agilent technologies, Santa Clara, CA, USA). For the gene ontology analyses, probe IDs were converted to unigene IDs, and unigenes were assigned gene ontology terms from the gene ontology database based on the closest gene ontology-annotated BLASTX homolog.
Primary breast cancer cell isolation from MMTV- PyMT mice
The tissues were minced into small (1–2 mm) pieces and digested with 5% FBS in DMEM containing 2 mg/ml collagenase I and 2 mg/ml hyaluronidase (Sigma-Aldrich) at 37 °C for 2 h. The cells were sequentially filtered through a 70-μm cell strainer and washed with PBS. The cells were then centrifuged in a Beckman Allegra X-15 R centrifuge at 300 × g for 10 min. The harvested cells were cultured in DMEM.
Mouse bone marrow transfer assay
Female FVB-MMTV-PyMT and Bcl-3−/− MMTV-PyMT mice (6–8 weeks old) were subjected to 8.5 Gy total body irradiation before being injected with 5 × 106 bone marrow cells isolated from female FVB mice in 0.2 ml of medium via the tail vein.
Statistical analyses
Unless stated otherwise, all experiments were repeated three times and all data were presented as mean±standard deviation. All other experiments were analyzed using two-tailed Student’s t-tests. P⩽0.05 was considered statistically significant.
References
Padua D, Massague J . Roles of TGFbeta in metastasis. Cell Res 2009; 19: 89–102.
Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E . Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol 2009; 11: 1287–1296.
Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008; 133: 66–77.
Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J . Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci USA 2003; 100: 8430–8435.
Thiery JP . Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2: 442–454.
Xi Q, He W, Zhang XH, Le HV, Massague J . Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. The J Biol Chem 2008; 283: 1146–1155.
Massague J, Seoane J, Wotton D . Smad transcription factors. Genes Dev 2005; 19: 2783–2810.
Liang YY, Brunicardi FC, Lin X . Smad3 mediates immediate early induction of Id1 by TGF-beta. Cell Res 2009; 19: 140–148.
Petersen M, Pardali E, van der Horst G, Cheung H, van den Hoogen C, van der Pluijm G et al. Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene 2010; 29: 1351–1361.
Al-Salihi MA, Herhaus L, Sapkota GP . Regulation of the transforming growth factor beta pathway by reversible ubiquitylation. Open Biol 2012; 2: 120082.
Bruce DL, Sapkota GP . Phosphatases in SMAD regulation. FEBS Lett 2012; 586: 1897–1905.
Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell 2009; 36: 457–468.
Herhaus L, Al-Salihi M, Macartney T, Weidlich S, Sapkota GP . OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun 2013; 4: 2519.
Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K et al. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol Biol Cell 2001; 12: 1431–1443.
Ohno H, Takimoto G, McKeithan TW . The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell 1990; 60: 991–997.
Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K et al. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell 1993; 72: 729–739.
Franzoso G, Bours V, Azarenko V, Park S, Tomita-Yamaguchi M, Kanno T et al. The oncoprotein Bcl-3 can facilitate NF-kappa B-mediated transactivation by removing inhibiting p50 homodimers from select kappa B sites. EMBO J 1993; 12: 3893–3901.
Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D . The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev 1993; 7: 1354–1363.
Franzoso G, Carlson L, Scharton-Kersten T, Shores EW, Epstein S, Grinberg A et al. Critical roles for the Bcl-3 oncoprotein in T cell-mediated immunity, splenic microarchitecture, and germinal center reactions. Immunity 1997; 6: 479–490.
Zhang X, Wang H, Claudio E, Brown K, Siebenlist U . A role for the IkappaB family member Bcl-3 in the control of central immunologic tolerance. Immunity 2007; 27: 438–452.
Wakefield A, Soukupova J, Montagne A, Ranger J, French R, Muller WJ et al. Bcl3 selectively promotes metastasis of ERBB2-driven mammary tumors. Cancer Res 2013; 73: 745–755.
Liu Z, Jiang Y, Hou Y, Hu Y, Cao X, Tao Y et al. The IkappaB family member Bcl-3 stabilizes c-Myc in colorectal cancer. J Mol Cell Biol 2013; 5: 280–282.
Zamora R, Espinosa M, Ceballos-Cancino G, Segura B, Maldonado V, Melendez-Zajgla J . Depletion of the oncoprotein Bcl-3 induces centrosome amplification and aneuploidy in cancer cells. Mol Cancer 2010; 9: 223.
Westerheide SD, Mayo MW, Anest V, Hanson JL, Baldwin AS Jr . The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G(1) transition. Mol Cell Biol 2001; 21: 8428–8436.
Ikeda F, Dikic I . CYLD in ubiquitin signaling and tumor pathogenesis. Cell 2006; 125: 643–645.
Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R . Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 2006; 125: 665–677.
Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T et al. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J Exp Med 2009; 206: 221–232.
Urban BC, Collard TJ, Eagle CJ, Southern SL, Greenhough A, Hamdollah-Zadeh M et al. BCL-3 expression promotes colorectal tumorigenesis through activation of AKT signalling. Gut 2015; 65: 151–164.
Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr . Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene 2000; 19: 1123–1131.
Pratt MA, Bishop TE, White D, Yasvinski G, Menard M, Niu MY et al. Estrogen withdrawal-induced NF-kappaB activity and bcl-3 expression in breast cancer cells: roles in growth and hormone independence. Mol Cell Biol 2003; 23: 6887–6900.
Cancer Genome Atlas Research Network Cancer Genome Atlas Research Network Weinstein JN, Cancer Genome Atlas Research Network Collisson EA, Cancer Genome Atlas Research Network Mills GB, Cancer Genome Atlas Research Network Shaw KR, Cancer Genome Atlas Research Network Ozenberger BA et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45: 1113–1120.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003; 3: 537–549.
Fantozzi A, Christofori G . Mouse models of breast cancer metastasis. Breast Cancer Res 2006; 8: 212.
DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009; 16: 91–102.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD et al. Genes that mediate breast cancer metastasis to lung. Nature 2005; 436: 518–524.
Drabsch Y, ten Dijke P . TGF-beta signaling in breast cancer cell invasion and bone metastasis. J Mammary Gland Biol 2011; 16: 97–108.
Derynck R, Zhang YE . Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003; 425: 577–584.
Zhang M, Wang M, Tan X, Li TF, Zhang YE, Chen D . Smad3 prevents beta-catenin degradation and facilitates beta-catenin nuclear translocation in chondrocytes. J Biol Chem 2010; 285: 8703–8710.
Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang XF . Axin and GSK3- control Smad3 protein stability and modulate TGF- signaling. Genes Dev 2008; 22: 106–120.
Blobe GC, Schiemann WP, Lodish HF . Role of transforming growth factor beta in human disease. N Engl J Med 2000; 342: 1350–1358.
Massague J . TGFbeta in Cancer. Cell 2008; 134: 215–230.
Matsuura I, Wang G, He D, Liu F . Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry 2005; 44: 12546–12553.
Remy I, Montmarquette A, Michnick SW . PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat Cell Biol 2004; 6: 358–365.
Chen N, Balasenthil S, Reuther J, Frayna A, Wang Y, Chandler DS et al. DEAR1 is a chromosome 1p35 tumor suppressor and master regulator of TGF-beta-driven epithelial-mesenchymal transition. Cancer DIscov 2013; 3: 1172–1189.
Conery AR, Cao Y, Thompson EA, Townsend CM Jr., Ko TC, Luo K . Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat Cell Biol 2004; 6: 366–372.
Padua D, Zhang XHF, Wang Q, Nadal C, Gerald WL, Gomis RR et al. TGFβ Primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008; 133: 66–77.
Schwarz EM, Krimpenfort P, Berns A, Verma IM . Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev 1997; 11: 187–197.
Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 2009; 325: 1134–1138.
Acknowledgements
This study was supported by grants from the National Basic Research Program (2014CB541904, 2014CB943600), the National Natural Science Foundation of China (31570902, 31370881). We thank Drs. F Liu for her gifts of antibodies, A Chariot and Y Zhang for their constructs, H Wei and Y Liu for their suggestions, and Q Jing for his support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, X., Cao, X., Sun, X. et al. Bcl-3 regulates TGFβ signaling by stabilizing Smad3 during breast cancer pulmonary metastasis. Cell Death Dis 7, e2508 (2016). https://doi.org/10.1038/cddis.2016.405
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2016.405
This article is cited by
-
Discovery of a small molecule that inhibits Bcl-3-mediated cyclin D1 expression in melanoma cells
BMC Cancer (2024)
-
Multifaceted roles for BCL3 in cancer: a proto-oncogene comes of age
Molecular Cancer (2024)
-
Multiple roles for Bcl-3 in mammary gland branching, stromal collagen invasion, involution and tumor pathology
Breast Cancer Research (2022)
-
Stanniocalcin 1 promotes metastasis, lipid metabolism and cisplatin chemoresistance via the FOXC2/ITGB6 signaling axis in ovarian cancer
Journal of Experimental & Clinical Cancer Research (2022)
-
BCL3 expression is strongly associated with the occurrence of breast cancer relapse under tamoxifen treatment in a retrospective cohort study
Virchows Archiv (2022)