Abstract
Melanoma inhibitory activity/cartilage-derived retinoic acid-sensitive protein (MIA/CD-RAP) is a small soluble protein secreted from chondrocytes. It was identified as the prototype of a family of extracellular proteins adopting an SH3 domain-like fold. In order to study the consequences of MIA/CD-RAP deficiency in detail we used mice with a targeted gene disruption of MIA/CD-RAP (MIA−/−) and analyzed cartilage organisation and differentiation in in vivo and in vitro models. Cartilage formation and regeneration was determined in models for osteoarthritis and fracture healing in vivo, in addition to in vitro studies using mesenchymal stem cells of MIA−/− mice. Interestingly, our data suggest enhanced chondrocytic regeneration in the MIA−/− mice, modulated by enhanced proliferation and delayed differentiation. Expression analysis of cartilage tissue derived from MIA−/− mice revealed strong downregulation of nuclear RNA-binding protein 54-kDa (p54nrb), a recently described modulator of Sox9 activity. In this study, we present p54nrb as a mediator of MIA/CD-RAP to promote chondrogenesis. Taken together, our data indicate that MIA/CD-RAP is required for differentiation in cartilage potentially by regulating signaling processes during differentiation.
Similar content being viewed by others
Main
Melanoma inhibitory activity/cartilage-derived retinoic acid-sensitive protein (MIA/CD-RAP) was cloned as a secreted protein from human melanoma cell lines1, 2 and independently identified by differential display comparing differentiated with dedifferentiated chondrocytes.3 Several studies indicate an important role of MIA/CD-RAP in tumor progression and metastasis of malignant melanoma by mediating cell detachment from extracellular matrix molecules such as fibronectin.4
In situ hybridization of mouse embryos revealed MIA/CD-RAP expression starting from the beginning of chondrogenesis and abundant throughout cartilage development.3, 5 Moreover, MIA/CD-RAP was revealed to be a specific marker for chondrocytic differentiation.6
Cartilage damage due to rheumatoid arthritis or mechanical overload, for instance during marathon running, releases MIA/CD-RAP from the chondroid matrix and can be monitored clinically by enhanced MIA/CD-RAP serum levels.7, 8 Based on its highly restricted activity, the MIA/CD-RAP promoter was used to study transcriptional mechanisms mediating chondrocyte differentiation. Expression of a LacZ-reporter gene under control of the 2251 bp MIA/CD-RAP promoter was exclusively observed in cartilage.9 These data suggested that MIA/CD-RAP may be functionally important for cartilage differentiation and for specific morphogenetic events during embryogenesis.
To study the consequences of MIA/CD-RAP deficiency in vivo we generated MIA/CD-RAP-deficient (MIA−/−) mice.10 First data gained from the analysis of the MIA−/− mice indicate that MIA/CD-RAP function is required for the highly ordered fibrillar ultra structure of cartilage. Electron-microscopic analyses reveal defects in collagen fiber density, diameter and arrangement as well as changes in the number and morphology of chondrocytic microvilli.10
In a recent study, we could show that MIA/CD-RAP can influence cartilage differentiation.11 MIA/CD-RAP was not able to induce chondrocytic differentiation of human mesenchymal stem cells on its own but induced differentiation mediated by transforming growth factor (TGF)-β3 and reverted osteogenic differentiation by BMP2 into more chondrogenic differentiation. Therefore, we aimed to analyse the molecular function of MIA/CD-RAP in cartilage differentiation using in vivo models for cartilage destruction and regeneration and in vitro models using mesenchymal stem cells of MIA−/− mice.
Results
In an initial study, we analyzed spontaneous emerging osteoarthritis (OA) in 2-years old mice and surprisingly revealed reduced OA in MIA−/− mice compared with WT (data not shown). We, therefore, aimed to analyze this finding in mouse models for cartilage destruction, regeneration and differentiation to reveal whether MIA is important for cartilage repair or cartilage resistance.
Models for osteoarthritis and fracture healing revealed enhanced cartilage repair in MIA−/− mice
To determine the effects of MIA/CD-RAP on cartilage under pathological situations we used MIA/CD-RAP-deficient mice in in vivo models for osteoarthritis and fracture healing. In both models cartilage degradation, regeneration and differentiation can be evaluated.
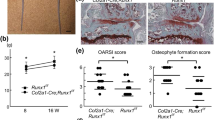
OA was induced in 6-week old mice by detaching the medial meniscotibial ligament from the tibia plateau and progression of OA was analyzed in mice knee joints after 10, 21 and 42 days. Interestingly, OA development scored by grade and stage was markedly reduced in MIA−/− mice compared with wild-type (WT) mice after 21 and 42 days (Figure 1a). Nevertheless, MIA−/− mice developed OA at all, which rose at day 21, but was remarkably reduced till day 42 pointing towards cartilage regeneration. Careful analyses of the medial part of knee joints in 42-days OA mice by hematoxylin and eosin (HE) staining revealed strong cartilage damage in WT mice in contrast to MIA−/− mice (Figure 1b). Surprisingly, proliferating chondrocytes, illustrated by PCNA staining in the knee joints of mice 10 days after OA was induced, were observed in MIA−/− mice but not in WT (Figure 1c). To analyse cartilage formation in MIA−/− mice in detail, we used the fracture model to observe de novo cartilage and bone formation in the callus. Bone formation was normal or even slightly reduced (Figure 1d). Interestingly, cartilage formation was significantly elevated in MIA−/− mice at day 7 after fracture. At day 9 and 11 the overall percentage of cartilage did not differ between WT and MIA−/−. To further evaluate cartilage differentiation and proliferation at day 7 after fracture, sections were stained with anti-collagen type II, type X and PCNA antibodies showing that the amount of proliferating collagen type II-positive undifferentiated chondrocytes was significantly enhanced in MIA−/− mice (Figure 1e). Hence, raised cartilage formation in MIA−/− was due to enhanced proliferation at early time points of fracture healing.
(a–c) Osteoarthritis (OA) was reduced in MIA−/− mice, mediated by enhanced proliferation. An experimental osteoarthritis model was used, in which instability to knee joints was created by detaching the medial meniscotibial ligament from the tibia plateau of 6-week old WT and MIA−/− mice. (a) After 10, 21 and 42 days OA development was scored by grade and stage. OA was markedly reduced in MIA−/− mice. We observed enhanced cartilage regeneration at day 42 in MIA−/− mice. The score of 10-days old control mice was zero. The data are represented as means±S.D. (b) HE staining of the medial part of knee joints 42 days after inducing osteoarthritis. Arrows indicate cartilage destruction. (c) PCNA immunhistological staining of knee joints, 10 days after OA was induced, showed PCNA positive chondrocytes in MIA−/− mice. Arrows indicate PCNA-positive cells. PCNA-positive cells in the knee joints were quantified. No PCNA-positive cells were found in WT mice. The data are represented as means±S.D. (d and e) Fracture healing model revealed enhanced cartilage formation. Tibia of 6–8-weeks old mice were broken and cartilage-/bone formation in the callus was analyzed after 5, 7, 9 and 11 days. (d) Cartilage formation was increased in MIA−/− mice compared with WT, whereas bone formation showed no difference. The data are represented as means±S.D. (e) Collagen type II, X and PCNA staining of serial sections of the callus 7 days after the fracture. PCNA staining revealed more proliferating collagen type II-positive cells in MIA−/− mice compared with control. Arrows present PCNA-positive cells. Small baskets indicate the augmented area, which displays the same detail of the respective serial section. (ns, not significant; *P<0.05; **P<0.01)
Accelerated proliferation and delayed differentiation in MIA−/− murine mesenchymal stem cells (mMSC)
As these models suggest accelerated proliferation of early chondrocytes before differentiation, we examined the effect of loss of MIA/CD-RAP on proliferation of primary mMSC derived from WT and MIA−/− littermates. Dynamic cell-cycle kinetic was analyzed in vitro through continued 5-bromo-2-deoxyuridine (BrdU) incubation using the flow cytometric BrdU/Hoechst quenching method in which up to three consecutive cell cycles can be investigated.
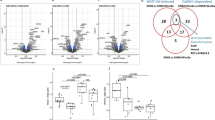
The effect of MIA/CD-RAP deficiency on cell-cycle progression in murine mesenchymal stem cells stained with Hoechst 33258 (Figure 2a, abscissa) and with Propidium iodide (PI) (Figure 2a, ordinate) is shown in Figure 2a after treatment with BrdU. After 96 h of BrdU incubation, more mMSC derived from MIA−/− mice had entered the first (42%) and second cell cycle (38%) compared with WT (first: 15%, second: 18%) (Figure 2b). A higher number of cells (66%) from WT littermates did not proliferate and represented G0/G1 cells of the first cell cycle than MIA/CD-RAP-deficient cells (20%). Comparing cell cycle distribution of proliferating cells between WT and MIA−/− mMSC, more MIA−/− stem cells were located in respective stages as WT (Figure 2b). In summary, cell-cycle progression was enhanced by loss of MIA/CD-RAP in murine mesenchymal stem cells.
Proliferation was enhanced in MIA−/− mMSC. (a) Modification of cell cycle distribution of murine mesenchymal stem cells (mMSC) by MIA/CD-RAP using BrdU/Hoechst quenching measurement. BrdU-quenched Hoechst33258 versus PI fluorescence density plots of cells after 96 h BrdU treatment. Cells of the first cell cycle are labeled G0/G1, S, and G2. Those of the second cycle are marked G1′, S′ and G2′. (b) Distribution of mMSC cells in cell cycle compartments. WT cells showed a delay in cell-cycle progression compared with MIA−/− cells represented by a high G0/G1 value of WT (66%) compared with MIA−/− (20%). In contrast to MIA−/− mMSC (80%) only 33% of all WT cells entered the first or second cell cycle after 96 h of incubation. The data are represented as means±S.D. (c) Comparison of the expression of Cyclin D2 using quantitative real-time PCR and western blot analysis in mMSC. Cyclin D2 expression was significantly increased in MIA−/− mMSC on mRNA and protein level (I). In each case two different isolations of untreated mMSC are shown in the western blot. WT mMSC transfected with siRNA specific for MIA/CD-RAP showed significant increase in Cyclin D2 expression (II). Re-expression of hMIA in MIA−/− mMSC led to significant decrease in Cyclin D2 mRNA expression (III). The data are represented as means±S.D., after normalization to WT mMSC or control. (d) Messenger RNA-expression analysis by qRT-PCR of murine mesenchymal stem cells transfected with siRNA specific for MIA/CD-RAP. MIA/CD-RAP was successfully downregulated compared with si control transfected cells. The data are represented as means±S.D., after normalization to the control siRNA. (e) Relative proliferation for 8 days compared between WT and MIA−/− mesenchymal stem cells using XTT assay. Differentiation was induced by TGF-β3. Induction of differentiation led to reduction of proliferation in WT cells (I), however, proliferation in MIA−/− cells as well as in siMIA-transfected cells stayed unchanged (III). MIA−/− cells in which hMIA was re-induced showed impaired proliferation after induction of differentiation with TGF-β3 (II). The data are represented as means±S.D., after normalization to untreated cells. Differentiation was diminished due to MIA/CD-RAP loss. (f) Col2 promoter activity, analyzed by reporter gene assay, was significantly decreased in MIA−/− mMSC (I) and in WT cells transfected with MIA/CD-RAP siRNA (II). Differentiation was induced by TGF-β3. Data are represented as means±S.D., after normalization to pGL2basic transfected cells. (ns, not significant; *P<0.05; **P<0.01; ***P<0.001)
RNA-array analysis of murine chondrocytes derived from MIA−/− and WT littermates revealed upregulation of Cyclin D2 in MIA−/− chondrocytes (unpublished data). Cyclin D2 (CCND2) gene is a member of type-D cyclin family, which binds to and activates cyclin-dependent kinase (CDK) 4/6.12 Activated CCND2-CDK4/6 complex phosphorylates downstream proteins, which are essential for cell-cycle progression from G1- to S-phase. Cyclin D2 is therefore an important initiator of proliferation.13
We determined differential Cyclin D2 mRNA expression by quantitative real-time PCR (qRT-PCR) and protein levels per western blot in chondrocyte progenitor cells. Murine mesenchymal stem cells from MIA−/− littermates were compared with WT. In-line with the gene-array analysis, the relative CCND2 mRNA and protein expression was significantly higher in MIA−/− MSC (Figure 2c I). Thus loss of MIA/CD-RAP resulted in increased CCND2 expression, which promotes enhanced proliferation.
To confirm these results we used siRNA against MIA/CD-RAP in WT mMSC and re-expressed human MIA (hMIA) in MIA−/− mMSC, respectively. The efficiency of knocking down MIA/CD-RAP was shown by qRT-PCR (Figure 2d). As anticipated, MIA/CD-RAP knockdown in WT cells resulted in overexpression of Cyclind D2 on mRNA and protein level (Figure 2c II). Re-expression of hMIA in MIA−/− mMSC led to a significant decrease in Cyclin D2 mRNA expression (Figure 2c III).
In order to examine the effect of MIA−/− on proliferation under chondrogenic conditions, proliferation of mesenchymal stem cells derived from WT and MIA−/− mice was analyzed in monolayer cultures by XTT assay over 8 days. Chondrogenic differentiation was induced by TGF-β3. TGF-β3 is implicated in the process of endochondral ossification, which is initiated by the inhibition of proliferation of mesenchymal stem cells and differentiation into chondrocytes.14 It, therefore, initiates a decline in proliferation of mesenchymal cells and promotes differentiation. In WT mMSC, relative proliferation was significantly reduced through treatment with TGF-β3 compared with MIA−/− mMSC where relative proliferation remained unchanged (Figure 2e I). Consistent with these results, re-expression of hMIA in MIA−/− cells re-established the WT phenotype (Figure 2e II). MIA/CD-RAP mRNA knockdown by siRNA in WT cells led to no alteration in proliferation after induction compared with control (Figure 2e III). These data suggest prolonged proliferation and possibly delayed differentiation because of lack of MIA/CD-RAP in chondrogenic conditions.
To show the impact of MIA/CD-RAP on cartilage differentiation on transcriptional level, transcriptional activation of the Col2a1 promoter, an early marker for cartilage differentiation,15 was analyzed in mMSC originated from WT and MIA−/− mice using a reporter gene assay. Differentiation was induced by TGF-β3. There was significant less activation of the Col2a1 promoter in MIA−/− compared with WT (Figure 2f I), and in WT cells in which MIA/CD-RAP was knocked down (Figure 2f II) suggesting that MIA/CD-RAP is an important factor for initiation of Col2a1 expression and differentiation.
Nuclear RNA-binding protein 54-kDa (p54nrb) is downregulated in cartilage of MIA−/− mice
In a study to identify MIA/CD-RAP regulated genes in malignant melanoma, a Powerblot analysis of MIA−/− cell clones of the melanoma cell line Hmb216 was performed. P54nrb was found to be a MIA/CD-RAP-regulated gene (Figure 3a). Alerted by a recent study presenting an important role of p54nrb in enhancing Sox9-mediated transcription of Col2a1,17 we concentrated on this MIA/CD-RAP target gene.
Strong downregulation of p54nrb in MIA−/− specimen. (a) Powerblot analysis of MIA/CD-RAP negative Hmb2-cells showed strong downregulation of p54nrb arrow indicates p54nrb. (b) Relative expression levels of p54nrb was significantly decreased in adult murine cartilage of ear and sternum of MIA−/− mice measured by qRT-PCR. Data are represented as means±S.D., after normalization to WT tissue. (c) P54nrb immunohistostaining of rips of 15.5-days old mouse embryos. Protein expression was reduced in MIA−/− mice. P54nrb expression was reduced during redifferentiation of dedifferentiated chondrocytes from MIA/CD-RAP deficient mice. (d) Relative mRNA expression levels of p54nrb were decreased in redifferentiated chondrocytes measured by qRT-PCR. Redifferentiation was induced by spheroid formation and treatment with TGF-β3. Data are represented as means±S.D., after normalization to untreated cells. (ns, not significant; *P<0.05; **P<0.01)
Expression analysis of p54nrb mRNA levels in cartilage of ear and sternum of adult mice (Figure 3b), as well as on protein levels in rips of 15.5-days old mouse embryos, using p54nrb-specific antibodies in immunohistochemistry (Figure 3c), revealed strong downregulation of p54nrb in MIA−/− mice. Next, we analyzed expression of p54nrb during redifferentiation of dedifferentiated chondrocytes in three-dimensional (3D) pellet cultures. We revealed strong induction of expression in WT chondrocytes, whereas p54nrb expression in MIA−/− cells was markedly reduced (Figure 3d).
P54nrb is a mediator of MIA/CD-RAP in chondrogenesis.
To confirm the functional role of MIA/CD-RAP and its downstream target p54nrb on differentiation and proliferation, siRNA technology against p54nrb was used. The efficiency of knocking down p54nrb was shown by qRT-PCR and western blot analysis (Figure 4a).
Enhanced proliferation and delayed differentiation in MIA−/− mMSC is mediated through p54nrb. (a) Western blot and mRNA expression analysis by qRT-PCR of murine mesenchymal stem cells transfected with siRNA specific for p54nrb. On protein and mRNA levels, p54nrb was successfully downregulated compared with si control transfected cells. The data are represented as means±S.D., after normalization to the control siRNA. (b) The transcriptional activity of the Col2a1 promoter (I) measured by reporter gene assay and Col2 mRNA expression levels (II) are reduced in WT cells transfected with siRNA against p54nrb. MIA−/− MSC showed no alteration in Col2a1 promoter activity (III) and Col2 mRNA expression (IV) after treatment with p54nrb siRNA. Simultaneous reexpression of hMIA and silencing of p54nrb led to decreased Col2a1 promoter activity. Cells transfected with control plasmid showed no significant difference in Col2a1 promoter activity (V). Differentiation was induced by TGF-β3. Data are represented as means±S.D., after normalization to pGL2basic transfected cells. (c) Overexpression of p54nrb in MIA−/− mMSC led to a significant decrease in Cyclin D2 mRNA expression (I). Relative expression of Cyclin D2 was increased through knockdown of p54nrb in WT mMSC (II). Data are represented as means±S.D., after normalization to control. (d) Relative proliferation for 8 days of MIA−/− and WT mesenchymal stem cells. Differentiation was induced by TGF-β3. MIA−/− cells in which hp54nrb was overexpressed showed significant reduction in proliferation when treated with TGF-β3 (I). WT cells in which p54nrb expression was knocked down by siRNA showed no alteration in relative proliferation (II). The data are represented as means±S.D., after normalization to untreated cells. Simultaneous reexpression of MIA/CD-RAP and knockdown of p54nrb in MIA−/− mMSC indicated no difference in relative proliferation, whereas reexpression of hMIA alone led to reduction of proliferation (III). Differentiation was induced by TGF-β3. The data are represented as means±S.D., after normalization to untreated cells. (ns, not significant; *P<0.05; **P<0.01; ***P<0.001)
Analysis of cartilage differentiation on transcriptional level measured by the activity of the Col2a1 promoter and Col2 mRNA levels showed a significant reduction in WT cells transfected with p54nrb siRNA compared with control (Figure 4b I and II). Interestingly, this result corresponded to the effects in MIA−/− MSC and in WT mMSC treated with siRNA against MIA/CD-RAP (Figure 2f). Col2a1 promoter activity and Col2 mRNA expression in MIA−/− MSC treated with siRNA against sip54nrb revealed no significant reduction of Col2a1 promoter activity as well as mRNA expression (Figure 4b III and IV). To confirm the regulation of Col2a1 by MIA/CD-RAP and p54nrb we simultaneously reexpressed hMIA and decreased p54nrb levels by siRNA technology in MIA−/− MSC. As expected, reexpression of MIA/CD-RAP led to an increase in Col2a1 promoter activity, whereas simultaneous knock down of p54nrb significantly diminished Col2a1 promoter activity (Figure 4b V). There was no significant difference in Col2a1 promoter activity in dependency on p54nrb in cells transfected with the control plasmid pCMX-PL1. Consequently, we could confirm that MIA/CD-RAP regulated Col2a1 expression through p54nrb.
Overexpression of human p54nrb (hp54nrb) in MIA−/− MSCs resulted in significant reduction of Cyclin D2 expression (Figure 4c I) and knockdown of p54nrb in MSC WT led to increased Cyclin D2 expression (Figure 4c II). In agreement with the data presented in Figure 2e II and III, MIA−/− cells transfected with hp54nrb showed reduced proliferation under treatment with TGF-β3 (Figure 4d I), whereas WT cells in which p54nrb expression was silenced showed no changes in relative proliferation under differentiating conditions (Figure 4d II). These data showed that MIA/CD-RAP and p54nrb have the same influence on proliferation and differentiation.
Reexpression of hMIA and simultaneous knockdown of p54nrb mRNA expression in MIA−/− mMSC resulted in no modification of proliferation despite induction of differentiation by TGF-β3 (Figure 4d III). P54nrb turned out to be the mediator of MIA/CD-RAP in promoting chondrogenesis by initiating inhibition of proliferation and inducing differentiation.
P54nrb is not regulated by ERK signalling
It is known that MIA/CD-RAP can inhibit ERK activity through negative regulation of integrin alpha 5 activity.18 In a recent study, inhibition of ERK signaling by MIA/CD-RAP could be confirmed in chondrocytes through binding to integrin alpha 5.11
Therefore, the role of ERK signaling on the regulation of p54nrb expression was analyzed. The ERK signalling pathway was specifically inhibited using the MEK1/2 inhibitors PD98059 and UO126 in WT MSC. The results revealed no regulation of p54nrb expression, neither on mRNA nor on protein level (Figure 5a).
(a) No regulation of p54nrb through ERK signaling. Inhibition of ERK signaling using MEK1/2 specific inhibitors UO126 and PD98059 in WT mMSC led to no regulation of p54nrb expression, neither on mRNA nor on protein level. The data are represented as means±S.D., after normalization to DMSO-treated cells. (b and c) Schematic drawing of MIA/CD-RAP regulation in cartilage development. MIA/CD-RAP mediates differentiation of mesenchymal stem cells through regulation of collagen type II expression through activation of p54nrb, arrows represent activation. Bold arrows indicate differentiation progress. (ns, not significant)
Discussion
It is known that MIA/CD-RAP is strongly induced at the beginning of chondrogenesis during fetal development3 and expressed throughout cartilage development.5 As MIA/CD-RAP was revealed to be a specific marker for chondrocytic differentiation,6 there is evidence for its role in cartilage development. Since the molecular function of MIA/CD-RAP in cartilage and bone formation was not analyzed in detail so far, we aimed to understand the role of MIA/CD-RAP in cartilage development. Previously, we could show that MIA/CD-RAP can modulate BMP2-induced osteogenic differentiation towards chondrogenic differentiation.19 In more detail, we could demonstrate inhibition of ERK signalling by MIA/CD-RAP, mediated through binding of MIA/CD-RAP to integrin alpha 5 and inhibiting its activity. Active ERK signalling is known to block chondrogenic differentiation, so MIA/CD-RAP stabilizes cartilage differentiation and inhibits differentiation into bone.11 Additionally, MIA/CD-RAP-deficient mice showed a disordered spatial arrangement of chondrocytes and an enlarged layer of calcified cartilage in articular cartilage indicating changes of cartilage maturation possibly involving an altered temporal pattern of proliferation.11 These results further support the hypothesis of MIA/CD-RAP being an important element in cartilage development.
As MIA−/− mice are viable, develop normally and histological examination of cartilage by means of light microscopy revealed no major abnormalities,10 we focused first on in vivo models, in which cartilage formation and regeneration could be studied. Here, we present in vivo models for fracture healing and osteoarthritis, which clearly showed enhanced chondrocytic regenerative capacity under MIA/CD-RAP deficiency. In a model for osteoarthritis, proliferating chondrocytes could be determined for the first time because of loss of MIA/CD-RAP clearly illustrating the highly regenerative potential in mature cartilage. Consequently, proliferating chondrocytes together with their increased matrix production not only counteract development of lesions in MIA−/− mice as seen at day 21 of OA compared with WT but also promotes enhanced regeneration at day 42, although OA proceeds in WT. The score of MIA−/− mice 42 days after OA induction decreased to the level of the control group, which leaves no doubt to the enhanced regenerative capacity by loss of MIA/CD-RAP. In fracture healing we revealed that cartilage formation proceeds faster in MIA−/− mice compared with WT through forced proliferation of collagen type II-positive undifferentiated chondrocytes under MIA/CD-RAP deficiency. However, as also seen in the adult MIA-deficient mice, the overall amount of cartilage did not differ hinting to secondary regulation counteracting lack of MIA/CD-RAP during development.
In cell-culture experiments we revealed that regeneration is enabled by the lack of MIA/CD-RAP through enhanced proliferation of mesenchymal stem cells and delayed differentiation during chondrogenesis. Enhanced proliferation is mediated by cell-cycle regulation through upregulation of Cyclin D2 expression promoting progression from G1 to S phase of cell cycle. Loss of MIA/CD-RAP decelerated differentiation exemplified by reduced Col2a1 promoter activity and by a delay in aggrecan mRNA expression (data not shown).
In this study, we could identify p54nrb as a mediator of MIA/CD-RAP in this regulation. Recent studies revealed that p54nrb physically interacts with Sox9 and enhances Sox9-dependent transcriptional activation of the Col2a1 promoter, which leads to progression of chondrogenesis.17 We were able to show that MIA/CD-RAP modulates chondrogenesis by initiating a stop of proliferation and inducing differentiation in both cases through p54nrb. P54nrb mediates differentiation through enhanced Sox9-dependent Col2a1 expression, whereas inhibition of proliferation is mediated by negative cell cycle regulation through Cyclin D2. The original findings that MIA/CD-RAP expression is regulated by the transcription factor Sox920 and AP-221 (Figure 5b) has to be modified in a way that MIA/CD-RAP can also act upstream of Sox9 to promote differentiation (Figure 5c). It is known that the MIA/CD-RAP promoter contains a SOX consensus sequence to which the Sox9 protein is able to bind, leading to a dose-dependent increased activity of the promoter.20 MIA/CD-RAP is specifically expressed during chondrogenesis5 and generally considered to be characteristic of cartilage.3 Surprisingly, Sox9 is not able to induce endogenous CD-RAP expression in mesenchymal cells in contrast to chondrocytes, which implies that other factors are involved in chondrogenesis,20 to induce the chondrocyte phenotype. This finding matches with our results, as we could show that in mesenchymal stem cells MIA/CD-RAP acts upstream from Sox9 and promotes chondrogenesis in early stages through p54nrb. We could reveal no regulation of p54nrb by MIA/CD-RAP through ERK-signalling and have to further analyse the regulation of p54nrb through MIA/CD-RAP.
In summary, our data indicate that MIA/CD-RAP is required for differentiation in cartilage upstream of the transcription factor Sox9. It regulates signalling processes during differentiation through p54nrb. Most impressively, the lack of MIA/CD-RAP expression results in cartilage repair and enhanced proliferation of cartilage precursor cells. We feel that this investigation of MIA/CD-RAP in cartilage will help to understand its molecular impact and will give new insight in the approach of developing therapies for cartilage regeneration in arthritis.
Materials and Methods
Mice
Transgenic animals were generated as described previously.10 C57Bl/6 control and MIA/CD-RAP-knockout mice were bred under specified pathogen-free conditions at 26°C, 70% relative humidity, and a 12-h light/12-h dark cycle at the University of Regensburg. They were fed with a breeding/maintenance diet (Altromin GmbH, Lage, Germany) and water ad libitum. The mice were randomly housed in polypropylene cages with sawdust bedding. The cages were sanitised twice weekly. Embryos were harvested on day 15.5 days post-coitum (dpc), whereas day 0.5 was on the day of detection of the vaginal plug. Animal care and all experimental procedures were carried out in accordance with guidelines of the German law governing animal care. Experiments were approved by the Ethics Committee for Animal Research of the Bavarian government.
Animal studies
Cartilage formation and regeneration was determined in models for osteoarthritis and fracture healing in vivo. Osteoarthritis in 6-week old mice (MIA−/− and WT) was induced by detaching the medial meniscotibial ligament from the tibia plateau. Briefly, under general anesthesia using intraperitoneal injections of Ketamin (90 mg kg−1) and Xylazin (10 mg/kg), the bilateral hind limbs were prepared for aseptic surgery. The knee joint was exposed following a medial incision without transection of the patellar ligament. Then, the medial meniscus was removed using a surgical microscope. After replacement of the extensor muscle, the medial incision was sutured, and the skin was closed. During the procedure, close attention was paid not to injure the articular cartilage. The mice were killed at day 10, and 3 and 6 weeks after operation, and the knees were dissected, fixed in 4% paraformaldehyd in phosphate-buffered saline (PBS), decalcified in 20% ethylene diamine tetraacetic acid (EDTA, Sigma, Taufkirchen, Germany), and paraffin embedded. Sections (5 μm) were cut, stained with HE, and the progress of OA was scored by stage and grade using the scoring system proposed by Pritzker et al.22 To analyse proliferating cells, PCNA staining was performed as described below.
For fracture healing, 6–8-weeks old mice (MIA−/− and WT) of both genders were used. After anesthesia with an intraperitoneal injection of Ketamin (90 mg kg−1) and Xylazin (10 mg kg−1) standardized non-stable fractures were produced distal to the crest of the tibia in accordance with the German animal protective law (Grant no. 54-2531.1-12/05). The mice were killed by CO2-intoxication at days 5, 7, 9 and 11 after fracture. The fractured tibia and the collateral, unfractured control tibiae were dissected, fixed in 4% freshly prepared paraformaldehyde in PBS, and paraffin embedded for immunohistological analysis. Sections (4 μm) were cut through the long axis of each tibia in a sagittal plane and adjacent sections were stained with Weigert's hematoxylin/alcian blue/sirius red (Sigma) or processed for immunohistochemistry. To evaluate the expression and localization of collagen type II, collagen type X and PCNA in the callus tissue during fracture healing, the following antibodies were used: mouse anti collagen type II (Developmental studies hybridoma bank, Iowa, USA), mouse anti collagen type X (Quartett, Berlin, Germany), and mouse anti PCNA (Dako, Hamburg, Germany). The deparaffinized and rehydrated serial sections were heat treated with 0.1 M citrate buffer, pH 6.0 in case of PCNA and preincubated with protease XXIV (Sigma; 0.05% in PBS for 10 min at 37°C) and testicular hyaluronidase (Sigma; 0.1% in acetate buffer, pH 6.0, for 90 min at 37°C) in case of collagen type II and X. After the pre-treatment, the sections were washed several times in PBS and briefly incubated with 0.3% H2O2 for 10 min at room temperature to block endogenous peroxidase activity. For the staining, a commercial biotinylation kit was used (ARK, Dako). The sections were counterstained in Weigert's hematoxylin, dehydrated and mounted. Sections incubated without the primary antibody served as a negative control. No sections showed any staining in such control experiments (data not shown). The evaluation of the staining was performed semiquantitatively by light microscopy. For histomorphometry, the callus proximally of the fracture line along the extensor side of the tibia was defined as the region of interest. To determine the cartilage and bone content in this region of interest, sagittal sections stained with Weiger's hematoxylin/alcian blue/sirius red from all time points and tibial growth plates from fractured and unfractured legs were photographed using a digital camera (DS-5M, Nikon, Düsseldorf, Germany). The images were imported into PhotoShop, and the software was used to select the areas of cartilage and bone in each callus. For the quantification of the selected pixels UTHSCSA ImageTool was used. The image software Eclipse-Net (Nikon) was used for the following histomorphometrical measurements: determination of the diameter of hypertrophic chondrocytes, and the quantification of hypertrophic chondrocytes per area.
Cell isolation and cell culture of primary murine MSCs and chondrocytes
MSCs were isolated from bone marrow of tibia and femur of 6–8-weeks old mice and grown in vitro in Dulbecco modified Eagle medium (DMEM) (PAA, Pasching, Austria), consisting of high glucose (Sigma), 20% fetal calf serum (FCS; PAN Biotech GmbH, Aidenbach, Germany), MEM vitamins (Invitrogen, Karlsruhe, Germany), penicillin (100 U per ml), streptomycin (10 μg ml−1) (both Sigma), and Amphotericin B (2.5 μg ml−1) (PAN Biotech GmbH) and were incubated in humidified atmosphere containing 5% CO2 at 37°C. Medium was replaced 24 h after isolation to select for plastic-adherent cells and every 3 days thereafter. Confluent cultures (∼3 × 104 cells per cm2) were passaged at 1 : 2. Cells were washed with PBS (pH 7.4, PAA) and were detached from culture flasks by incubating for 3 min at 37°C in a PBS solution supplemented with 0.05% trypsin and 0.02% ethylenediaminetetraacetic acid (Invitrogen). All experiments were performed at passage three to six.
MSC were treated with DMSO, UO126 and PD098059, respectively, (all CALBIOCHEM, Nottingham, UK) and incubated for 48 h followed by RNA and protein isolation.
BrdU/Hoe-quenching technique
For BrdU/Hoechst33258 (Hoe)-quenching measurements, 5 × 105 cells were seeded on day 0 into T25 culture flasks (Corning Inc., Corning, NY, USA) and were incubated for 7 days. On day 3 and 5 after cell seeding, medium was changed and cells were treated with 120 μM BrdU (Sigma). To minimize potential disturbance in the nucleotide pathway due to BrdU treatment, medium was also supplemented with half-equimolar 2′-deoxycytidine (Sigma).23 For BrdU/Hoe-quenching cell staining, cells were harvested on day 7 through trypsination and washed twice with ice cold DNA-washing buffer (100 mM Tris-HCl pH 7.4, 154 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.2% bovine serum albumin (BSA)) (all Merck, Darmstadt, Germany). A total of 5 × 105 cells were resuspended in 0.5 ml DNA-staining buffer (100 mM Tris-HCl, pH 7.4, 154 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.2% BSA (all Merck), 0.1% IGEPAL CA-630 (Nonylphenyl-polyethylenglycol) (Sigma) supplemented with 40 μg ml−1 (2–4 U per ml) RNase (Roche Applied Science, Mannheim, Germany) and 1.2 μg ml−1 Hoechst 33258 (Sigma) and incubated for 15 min at 37°C.24, 25 Cellular DNA content was stained with PI (1.5 μg ml−1) (Sigma) for 15 min on ice.
Flow cytometry instrumentation and data analysis
An LSR-I three laser flow cytometer (Becton Dickinson Biosciences, San Jose, CA, USA) was used for BrdU/Hoe-quenching measurements. A 325 nm ultraviolet laser line was used for Hoe excitation. PI fluorescence was excited with an air-cooled 488 nm argon laser. Fluorescence emission was detected through 425/44 band-pass and 630/22 band-pass filter optic, respectively. Sample measurements and data analysis were performed with Diva software 6.1.1 (Becton Dickinson Biosciences). Cell debris and aggregates were excluded from analysis using pulse processing (DNA width signal against DNA area signal). By drawing regions around the initial G0 cell fraction (Figure 3a), we identified cells that remained BrdU negative within a period of 96 h of BrdU incubation time, indicating quiescent cells. Quantification of cell cycle distribution was performed with FlowJo software (Tree Star Inc., Ashland, OR, USA).
siRNA knockdown
MSCs were transfected with gene-specific siRNA against p54nrb purchased from Qiagen (Hilden, Germany) or MIA/CD-RAP purchased from Sigma. A negative scramble siRNA (Qiagen) was used as a control. Transfection was performed according to the manufacturer's specification using Lipofectamine 2000 reagent (Invitrogen). At 2 days after transfection, cells were harvested for RNA isolation and plated for proliferation assays as described elsewhere. For Luciferase assay cells were used after 1 day of transfection as described below.
Nucleofection with plasmid DNA
Nucleofection of mMSC was performed using the Human MSC Nucleofector Kit from Lonza (Basel, Switzerland). Manufacturer's instructions have been slightly modified. A total of 1 × 106 cells were resuspended in 100 μl complete nucleofector solution. The pCMX-PL1-human MIA/CD-RAP plasmid or the pCMV-SPORT6-hp54nrb plasmid (ImaGenes, Berlin, Germany, BC028299) both under the regulation of a cytomegalovirus (CMV) enhancer/promoter element, was then added (2 μg per transfection sample) to the cell suspension. As a control, cells received the empty expression construct pCMX–PL1. The cell/DNA mixtures were transferred into certified cuvettes (Lonza) and transfected by using program C-17. Following the electroporation, the cells were incubated in culture medium pre-heated to 37°C for 2 days, and then seeded for proliferation assays, luciferase assay and RNA harvest as described elsewhere.
Differentiation
For analysis of chondrogenesis murine MSC differentiation and chondrocyte redifferentiation was performed as 3D pellets. The 3D pellet culture facilitates the in vitro differentiation of mesenchymal stem cells into chondrocytes.26 For this, 2.5 × 105 cells were seeded into 15 ml polypropylene tubes (Greiner Bio-One GmbH, Frickenhausen, Germany) and pelleted at 150 r.c.f. for 2 min. Cells were cultured for the indicated period as 3D pellets in induction medium including DMEM (PAA), high glucose (Sigma), 20% FCS (PAN Biotech GmbH), MEM Vitamins (Invitrogen), penicillin (100 U/ml), streptomycin (10 μg/ml) (both Sigma), Amphotericin B (2.5 μg/ml) (PAN Biotech GmbH), 0.1 μM dexamethasone, 1 mM sodium pyruvate, 0.17 mM ascorbic acid-2-phosphate, 0.35 mM proline (all Sigma), insulin (5 μg/ml), transferring (5 μg), selenious acid (5 ng) (ITS Premix, Becton Dickinson, Minneapolis, MN, USA) and 10 ng/ml human TGF-β3 (R&D Systems). The medium was changed every second day. All groups were done in triplicate. Each experiment was repeated three times. RNA was harvested at days 2, 4 and 6 in case of mMSC and every day between 1 and 6 days in case of chondrocytes.
RNA isolation and RT-PCR
Total RNA from cell culture, tissue (sternum and ear of 6–8-weeks old mice) or 3D pellets was isolated using e.Z.N.A. MicroElute Total RNA Kit (peqlab Biotechnologie GmbH, Erlangen, Germany) as described by the manufacturers. Each three mMSC pellets were pooled and total RNA was isolated after indicated days of culture. RNA concentration and purity was measured by NanoDrop (peqlab Biotechnologie GmbH). cDNA was generated by RT. The RT reaction was performed in 20 μl reaction volume containing 500 ng of total RNA, 4 μl of 5 × first-strand buffer, 2 μl of 0.1 M DTT (both Invitrogen), 1 μl of dN6 primer (10 mM) (Roche, Mannheim, Germany), 1 μl of dNTPs (10 mM) (Amersham pharmacia biotech, Pittsburgh, PA, USA) and DEPC water. The reaction mix was incubated for 10 min at 70°C. Then 1 μl of Superscript II reverse transcriptase (Invitrogen) was added, and RNA was transcribed for 1 h at 37°C. Subsequently, reverse transcriptase was inactivated at 70°C for 10 min, and RNA was degraded by digestion with 1 μl RNase A (10 mg/ml) (Roche) at 37°C for 30 min. The quality of cDNA was controlled and normalized by qRT-PCR amplification of the house keeping gene β-actin.
Quantitative real-time PCR
Quantitative RT-PCR was performed with a Lightcycler (Roche). A volume of 2 μl cDNA template, 0.5 μl of forward and reverse primer (20 μM) for each collagen type II, p54nrb or Cyclin D2 and 10 μl of SYBR Premix Ex Taq (TaKaRa, Otsu, Japan) were combined to a total volume of 20 μl. PCR primers were obtained from Sigma.
The PCR program used was 30 s, 95°C (initial denaturation); 20°C s−1 temperature transition rate upto 95°C for 3 s; 10 s 58°C; 8 s 72°C, 85°C acquisition mode single, repeated for 40 times (amplification). Annealing and acquisition temperatures were optimized for each primer set. The PCR product was evaluated by melting-curve analysis according to the manufacturer's instructions and checking the PCR products on 1.8% agarose gels. Each analysis was performed at least in triplicate. The expression ratios of the analyzed genes were calculated using an internal control standard curve of β-actin levels. Primers used for PCR are shown in Table 1.
Luciferase assay
For transient transfections, 2 × 105 mMSC were seeded into each well of a six-well plate and transfected with 0.5 μg plasmid DNA using the Lipofectamine plus method (Gibco, Karlsruhe, Germany) according to the manufacturer's instruction. Cells were cultured in induction medium (see above) including 10 ng/ml human TGF-β3 (R&D Systems) to induce chondrogenic differentiation. The cells were lysed 24 h after transfection and the luciferase activity in the lysate was quantified by a luminometric assay (Promega Corp., Madison, WI, USA). Transfection efficiency was normalized according to renilla luciferase activity by cotransfecting 0.1 μg of the plasmid pRL-TK (Promega, Mannheim, Germany). All transfections were repeated at least three times. For transient transfection the COL2A1 LUC reporter construct27 kindly provided by Linda J. Sandell was used.
Cell proliferation kit II (XTT)
Cells were seeded as monolayer cultures in 100 μl cultivation medium (see above) without phenol red in a 96-well plate (Corning Inc.) at a density of 1 × 104 and 2 × 104 cells per well and incubated in humidified atmosphere containing 5% CO2 at 37°C for the indicated time periods. Experiments were performed in triplicates. Medium was changed after 4 days. Differentiation was induced by 10 ng/ml TGF-β3 (R&D Systems). After 2, 4, 6 and 8 days of incubation, 50 μl of XTT labeling and electron-coupling reagent (Roche) was added to the respective well, and plates were incubated at 37°C. Absorbance was measured after 2 and 4 h of incubation at 450 nm against a reference wavelength at 650 nm using a microplate reader (Molecular Devices, Sunnyvale, CA, USA, Emax precision microplate reader).
Western blotting
Cells were lysed in 75 μl RIPA buffer (Roche) and incubated for 15 min at 4°C. Insoluble fragments were removed by centrifugation at 16 100 r.c.f. for 10 min at 4°C and the supernatant was stored at −20°C. Protein concentrations were determined using the BCA protein assay reagent (Pierce, Rockford, IL, USA). In all, 75 μg (Cyclin D2) and 50 μg (p54nrb) protein per lane were denatured at 70°C for 10 min after addition of Roti-load-buffer (Roth, Karlsruhe, Germany) and subsequently separated on 12–15% SDS-PAGE gels. After blotting onto PVDF-membranes (Bio-Rad, Richmond, CA, USA) and blocking for 1 h with 3% BSA/PBS (p54nrb) and 5% milk/TBST (0.1%) (Cyclin D2), respectively, membranes were incubated overnight at 4°C with one of the following antibodies: anti-p54nrb (BD Biosciences, Bedford, MA, USA), anti-CyclinD2 (Abcam, Cambridge, UK), and anti β-actin (Sigma, Deisenhofen, Germany). After three washing steps with PBS and TBST (0.1%), respectively, the membrane was incubated for 1 h with an alkaline phosphate-coupled secondary anti-mouse IgG antibody (Chemicon, Hofheim, Germany) and then washed again. Finally, immunoreactions were visualized by BCIP/NBT (Sigma) staining.
Immunohistochemistry
Paraffin-embedded preparations of 15.5-days old WT and MIA−/− embryos were screened for p54nrb protein expression by immunohistochemistry. Samples were deparaffinised, rehydrated, heat treated with Tris–EDTA and subsequently incubated with primary monoclonal mouse anti p54nrb antibody (1 : 10000; BD Biosciences) for 30 min at room temperature. The secondary antibody Envision (Dako) was incubated for 30 min at room temperature. Antibody binding was visualized using DAB-Chromogen (Dako). Finally, the tissues were counterstained by hematoxylin. The evaluation of the staining was performed semi quantitatively by light microscopy.
PowerBlot
We used a PowerBlot Western Array screening system conducted by BD Biosciences to reveal regulated proteins in HMB2 cell clones stable transfected with a MIA/CD-RAP-antisense expression plasmid published by Tatzel et al.16 Experiments were conducted in triplicates.
Plasmids
The expression vectors used for re-expression were pCMX-PL1-human MIA/CD-RAP and pCMV-SPORT6-hp54nrb containing full-length cds cloned by ImaGenes. The sequence of the hp54nrb construct was confirmed by DNA sequencing performed at Entelechon, Regensburg, Germany.
Statistical analysis
The mean and S.D. values were calculated from independent experiments, and the graphs were constructed with the GraphPad Prism software. Statistical significance between two groups was determined by using the Student's t-test. Software used was GraphPad Prism 4. Statistical significance was analyzed on data from at least three independent experiments. A P-value <0.05 was considered statistically significant (NS, not significant; *P<0.05; **P<0.01; ***P<0.001).
Abbreviations
- BrdU:
-
5-bromo-2-deoxyuridine
- BSA:
-
bovine serum albumin
- DTT:
-
dithiothreitol
- EDTA:
-
ethylene diamine tetraacetic acid
- HE:
-
hematoxylin and eosin
- MIA/CD-RAP:
-
melanoma inhibitory activity/cartilage-derived retinoic acid-sensitive protein
- mMSC:
-
murine mesenchymal stem cells
- OA:
-
osteoarthritis
- p54nrb:
-
nuclear RNA-binding protein 54-kDa
- PBS:
-
phosphate-buffered saline
- qRT-PCR:
-
quantitative reverse transcription-polymerase chain reaction
- TGF:
-
transforming growth factor
References
Blesch A, Bosserhoff AK, Apfel R, Behl C, Hessdoerfer B, Schmitt A et al. Cloning of a novel malignant melanoma-derived growth-regulatory protein, MIA. Cancer Res 1994; 54: 5695–5701.
van Groningen JJ, Bloemers HP, Swart GW . Identification of melanoma inhibitory activity and other differentially expressed messenger RNAs in human melanoma cell lines with different metastatic capacity by messenger RNA differential display. Cancer Res 1995; 55: 6237–6243.
Dietz UH, Sandell LJ . Cloning of a retinoic acid-sensitive mRNA expressed in cartilage and during chondrogenesis. J Biol Chem 1996; 271: 3311–3316.
Bosserhoff AK, Stoll R, Sleeman JP, Bataille F, Buettner R, Holak TA . Active detachment involves inhibition of cell-matrix contacts of malignant melanoma cells by secretion of melanoma inhibitory activity. Lab Invest 2003; 83: 1583–1594.
Bosserhoff AK, Kondo S, Moser M, Dietz UH, Copeland NG, Gilbert DJ et al. Mouse CD-RAP/MIA gene: structure, chromosomal localization, and expression in cartilage and chondrosarcoma. Dev Dyn 1997; 208: 516–525.
Bosserhoff AK, Buettner R . Establishing the protein MIA (melanoma inhibitory activity) as a marker for chondrocyte differentiation. Biomaterials 2003; 24: 3229–3234.
Muller-Ladner U, Bosserhoff AK, Dreher K, Hein R, Neidhart M, Gay S et al. MIA (melanoma inhibitory activity): a potential serum marker for rheumatoid arthritis. Rheumatology (Oxford) 1999; 38: 148–154.
Neidhart M, Muller-Ladner U, Frey W, Bosserhoff AK, Colombani PC, Frey-Rindova P et al. Increased serum levels of non-collagenous matrix proteins (cartilage oligomeric matrix protein and melanoma inhibitory activity) in marathon runners. Osteoarthritis Cartilage 2000; 8: 222–229.
Xie WF, Zhang X, Sandell LJ . The 2.2-kb promoter of cartilage-derived retinoic acid-sensitive protein controls gene expression in cartilage and embryonic mammary buds of transgenic mice. Matrix Biol 2000; 19: 501–509.
Moser M, Bosserhoff AK, Hunziker EB, Sandell L, Fassler R, Buettner R . Ultrastructural cartilage abnormalities in MIA/CD-RAP-deficient mice. Mol Cell Biol 2002; 22: 1438–1445.
Schubert T, Schlegel J, Schmid R, Opolka A, Grassel S, Humphries M et al. Modulation of cartilage differentiation by MIA/CD-RAP. Exp Mol Med 2010; 42: 166–174.
Tashiro E, Tsuchiya A, Imoto M . Functions of cyclin D1 as an oncogene and regulation of cyclin D1 expression. Cancer Sci 2007; 98: 629–635.
Kobayashi T, Nakamura E, Shimizu Y, Terada N, Maeno A, Kobori G et al. Restoration of cyclin D2 has an inhibitory potential on the proliferation of LNCaP cells. Biochem Biophys Res Commun 2009; 387: 196–201.
Jin EJ, Choi YA, Sonn JK, Kang SS . Suppression of ADAM 10-induced Delta-1 shedding inhibits cell proliferation during the chondro-inhibitory action of TGF-beta3. Mol Cells 2007; 24: 139–147.
Lefebvre V, Smits P . Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today 2005; 75: 200–212.
Tatzel J, Poser I, Schroeder J, Bosserhoff AK . Inhibition of melanoma inhibitory activity (MIA) expression in melanoma cells leads to molecular and phenotypic changes. Pigment Cell Res 2005; 18: 92–101.
Hata K, Nishimura R, Muramatsu S, Matsuda A, Matsubara T, Amano K et al. Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J Clin Invest 2008; 118: 3098–3108.
Bauer R, Humphries M, Fassler R, Winklmeier A, Craig SE, Bosserhoff AK . Regulation of integrin activity by MIA. J Biol Chem 2006; 281: 11669–11677.
Tscheudschilsuren G, Bosserhoff AK, Schlegel J, Vollmer D, Anton A, Alt V et al. Regulation of mesenchymal stem cell and chondrocyte differentiation by MIA. Exp Cell Res 2006; 312: 63–72.
Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ . Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res 1999; 14: 757–763.
Xie WF, Kondo S, Sandell LJ . Regulation of the mouse cartilage-derived retinoic acid-sensitive protein gene by the transcription factor AP-2. J Biol Chem 1998; 273: 5026–5032.
Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage 2006; 14: 13–29.
Diermeier S, Schmidt-Bruecken E, Kubbies M, Kunz-Schughart LA, Brockhoff G . Exposure to continuous bromodeoxyuridine (BrdU) differentially affects cell cycle progression of human breast and bladder cancer cell lines. Cell Prolif 2004; 37: 195–206.
Poot M, Hoehn H, Kubbies M, Grossmann A, Chen Y, Rabinovitch PS . Cell-cycle analysis using continuous bromodeoxyuridine labeling and Hoechst 33358-ethidium bromide bivariate flow cytometry. Methods Cell Biol 1994; 41: 327–340.
Kubbies M . High-resolution cell cycle analysis: the flow cytometric bromodeoxyuridine-Hoechst quenching technique. In: Radbruch A (ed). Flow Cytometry and Cell Sorting, 2nd edn. 1999.
Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU . In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res 1998; 238: 265–272.
Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ et al. Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1beta. J Biol Chem 2003; 278: 17688–17700.
Acknowledgements
We thank Sibylla Lodermeyer and Elisabeth Schmidt-Bruecken for excellent technical assistance, PD Dr. Gero Brockhoff for perfect support in Flow cytometry and Linda Sandell for supplying the Col2a1 luciferase construct. This work was supported by grants from the Deutsche Forschungsgemeinschaft to AKB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Piacentini
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Schmid, R., Schiffner, S., Opolka, A. et al. Enhanced cartilage regeneration in MIA/CD-RAP deficient mice. Cell Death Dis 1, e97 (2010). https://doi.org/10.1038/cddis.2010.78
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2010.78
Keywords
This article is cited by
-
Activating enhancer binding protein 2 epsilon (AP-2ε)-deficient mice exhibit increased matrix metalloproteinase 13 expression and progressive osteoarthritis development
Arthritis Research & Therapy (2015)
-
SOX10 Promotes Melanoma Cell Invasion by Regulating Melanoma Inhibitory Activity
Journal of Investigative Dermatology (2014)
-
MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression
Oncogene (2013)
-
Post-traumatic osteoarthritis: from mouse models to clinical trials
Nature Reviews Rheumatology (2013)
-
Analysis of apoptosis methods recently used in Cancer Research and Cell Death & Disease publications
Cell Death & Disease (2012)