Abstract
Fas ligand (FasL) is a transmembrane protein that regulates cell death in Fas-bearing cells. FasL-mediated cell death is essential for immune system homeostasis and the elimination of viral or transformed cells. Because of its potent cytotoxic activity, FasL expression at the cell surface is tightly regulated, for example, via processing by ADAM10 and SPPL2a generating soluble FasL and the intracellular fragments APL (ADAM10-processed FasL form) and SPA (SPPL2a-processed APL). In this study, we report that FasL processing by ADAM10 counteracts Fas-mediated cell death and is strictly regulated by membrane localization, interactions and modifications of FasL. According to our observations, FasL processing occurs preferentially within cholesterol and sphingolipid-rich nanodomains (rafts) where efficient Fas–FasL contact occurs, Fas receptor and FasL interaction is also required for efficient FasL processing, and FasL palmitoylation, which occurs within its transmembrane domain, is critical for efficient FasL-mediated killing and FasL processing.
Similar content being viewed by others
Main
FasL (Fas ligand) is a transmembrane protein belonging to the TNF family, which, upon binding to the Fas receptor, mediates cell death in Fas-expressing cells.1 In vivo, FasL expression is mainly detected in hematopoietic cells where it has a key role in the induction of cell death by mediating the activation-induced cell death of T lymphocytes, therefore contributing to the cytotoxic effector function of T and NK cells.2 Moreover, FasL signaling has been reported to be critical in both the maintenance of immune privilege and the establishment of tumor survival.3, 4 The latter role has prompted the establishment of the controversial ‘tumor counterattack’ model, which suggests that FasL expression by tumor cells would kill Fas-sensitive effector cells of the immune system.5, 6
In addition to its pro-apoptotic activity, FasL has been implicated in retrograde signal transduction within FasL-expressing cells, also called reverse signaling, which is thought to be important in the modulation of T-cell activation. Humans and mice, with deleterious mutations in Fas or FasL, develop a severe lymphoaccumulative disease with an accumulation of aberrant T-cell population, leading to lymphadenopathy, splenomegaly and lupus erythematosus.
While Fas is expressed by several tissues, the expression of FasL is tightly regulated at both transcriptional and post-transcriptional levels to avoid undesirable cell death. The latter include FasL storage in intracellular compartments,7 recruitment into cholesterol- and sphingolipid-rich nanodomains (rafts),8, 9 and FasL proteolysis by matrix metalloproteinases (MMPs) and the intramembrane protease SPPL2a.10, 11
We and others have previously reported that the sequential processing of FasL by ADAM10 and SPPL2a generates soluble extracellular FasL (sFasL), a membrane-anchored 17 kDa APL fragment10, 11 and the intracellular 11 kDa SPPL2a cleavage product SPA.10 Although several reports have suggested that sFasL is less cytotoxic than the membrane-bound form of FasL,12, 13 a recent study clearly demonstrated in vivo that only the membrane-anchored FasL can efficiently induce cell death,14 thus highlighting the importance of FasL processing by ADAM10 and SPPL2a in the control of Fas-mediated cell death. Nevertheless, the molecular events that regulate FasL proteolysis have not been addressed.
We have demonstrated that the partitioning of both FasL and Fas into rafts is necessary for efficient cell death transduction. However, both the regulation of FasL targeting into the rafts and the importance of this localization for FasL proteolytic processing remain unclear.
In the present study, we show that processing of hFasL by ADAM10, but not by SPPL2a, counteracts FasL-mediated apoptosis. Confinement of proteins into rafts can be mediated by palmitoylation, that is, the reversible addition of a palmitate moiety into cysteine residues. In this study, we establish that hFasL, like its receptor Fas, is palmitoylated. More importantly, we demonstrate that partitioning inside rafts, the interaction with Fas receptor and palmitoylation are all essential events for hFasL proteolysis and function.
Results
hFasL processing by metalloproteases, but not by SPPL2a, reduces hFasL-induced cell death
We first aimed at studying the importance of sequential hFasL shedding for the pro-apoptotic function of hFasL. For this purpose, hFasL cleavage in WSU B cells stably transfected with hFasL was prevented by using the SPPL2a inhibitor (Z-LL)2 or the MMP inhibitor TAPI-2, and cells were co-cultured with Fas-expressing JH6.2 Jurkat target cells. We found that the killing capacity of hFasL was unaffected upon inhibition of the intramembrane protease SPPL2a by (Z-LL)2 (Figure 1a), but was increased after inhibition of metalloproteases (Figure 1b). Sufficient inhibition of hFasL cleavage was confirmed by western blot analysis of lysates prepared from (Z-LL)2- and TAPI-2-treated WSU hFasL cells. These data indicate that Fas-mediated cell death is regulated by ADAM10-processing of hFasL, which may function to hinder the pro-apoptotic function of hFasL.
Metalloprotease, but not SPPL2a-mediated, FasL processing decreases FasL-induced cell death. WSU cells were pre-treated with inhibitors of either MMP or SPPL2a before co-culture with the Fas-bearing JH6.2 cells (inhibitors remained in media during the co-culture period). Effective inhibition of ADAM10 or SPPL2 was confirmed by western blot. G247 antibody was used to detect the full-length FasL and the antibody Ab-3 was employed for the detection of the N-terminal APL and SPA fragments, ezrin was used as loading control. Cell death was quantified by flow cytometric quantification of the subG1 population of propidium iodide-stained ethanol-fixed cells. (a) JH6.2 cells were co-cultured for 6 hours with WSU cells stably transfected with either hFasL or mock transfected, and were either pre-treated with the SPPL2 inhibitor (Z-LL)2 (2 μM, 2 h) or left untreated. The graph represents the average of two independent experiments, with error bars indicating the S.D. (b) JH6.2 cells were co-cultured for 4 h with WSU cells stably transfected with hFasL or mock transfected and either pre-treated with the MMP inhibitor TAPI-2 (50 μM, 2 h) or left untreated. The graph represents the average of four independent experiments, with error bars indicating the S.D.
Optimal processing by ADAM10 requires hFasL localization into rafts and binding to the Fas receptor
To test the consequences of raft localization on hFasL processing, we performed a cholesterol oxidase treatment, which partially inhibits lipid nanodomain formation by converting cholesterol to cholestenone. The distribution of full-length FasL and APL in rafts were then analyzed using a biochemical approach. Post-nuclear supernatants (PNSs) were solubilized by polyoxyethylene ether Brij 98, and lysates were subjected to ultracentrifugation through a sucrose gradient. The various fractions were analyzed by western blotting. The membrane was probed with antibodies recognizing the raft protein marker Fyn and the non-raft marker Rab5a, as a control for proper separation. Our data show that whereas full-length FasL is only partially located in the raft fractions, the APL fragment was exclusively found there. In addition, the cholesterol oxidase treatment decreased the shedding of hFasL, thereby indicating that raft localization has a crucial role in ADAM10 processing of hFasL (Figure 2a).
hFasL processing depends on the lipid nanodomain integrity and on the binding to Fas receptor. (a) Stable WSU-hFasL cells were treated with cholesterol oxidase (CO; 4 U/ml) for 4 h or left untreated (NT). Cells were then solubilized in Brij 98 detergent and subjected to sucrose gradient separation before analysis of the heavy fraction (HF) and the light fraction (LF) by western blot analysis. (b) Cell lysates of WSU cells stably transfected with wild-type hFasL or hFasLA247E were analyzed by western blotting. G247 antibody was used to detect the full-length FasL and the antibody Ab-3 was used to measure the N-terminal APL fragment; Ezrin was used as loading control. (c) Stable WSU hFasL cells were treated with the metalloprotease inhibitors TAPI-2 (50 μM), GI254023x (10 μM) or INCB-3619 (10 μM) for 4 h or left untreated (NT). Cells were then solubilized in Brij 98 detergent and subjected to sucrose gradient separation before western blot analysis of the heavy and light fractions (identified respectively by Rab5 and Fyn). G247 antibody was used to detect the full-length FasL and the antibody Ab-3 was used to detect the N-terminal APL fragment. Ezrin was used as loading control
We also aimed at determining whether binding of hFasL to the Fas receptor is required for hFasL cleavage. The point mutation A247E in the Fas receptor-binding domain prevents interaction of hFasL with its receptor thereby preventing cell-mediated apoptosis (Supplementary Figure 1A and Del-Rey et al.24). Lysates prepared from hFasLA247E-transfected WSU cells showed decreased levels of the APL fragment compared with cells transfected with wild-type hFasL (Figure 2b). Similarly, transient transfection of Fas receptor targeting siRNA in HEK293 cells dramatically decreased the amount of APL (Supplementary Figure 1B). Collectively, these data demonstrate that binding to the Fas receptor is required for the processing of hFasL by ADAM10.
ADAM10 processing of hFasL occurs within rafts
It has been shown that differential nanodomain localization of target protein and metalloprotease can regulate the proteolytic processing.18, 19 Moreover, the above results indicate that hFasL processing mainly occurs within rafts. To obtain further evidence for this hypothesis, we treated hFasL-transfected WSU cells with the metalloprotease inhibitors TAPI-2, GI254023X or INCB-3619. Biochemical raft analysis showed that inhibition of ADAM10-mediated proteolysis of hFasL decreased APL production, while simultaneously increased the amount of full-length FasL preferentially inside the raft fractions (Figure 2c). Thus, our results support the notion that hFasL proteolysis by ADAM10 occurs within raft nanodomains.
Palmitoylation of hFasL is required for efficient processing by ADAM10 and for FasL-induced cell death
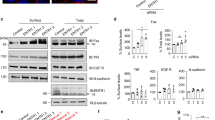
Protein palmitoylation represents one of the raft targeting signals. Using acyl biotinyl-exchange chemistry, we show that hFasL is indeed palmitoylated. Incorporation of point-mutated hFasL revealed that this post-translational modification occurs at the cysteine position 82, which is located at the N-terminal portion of the transmembrane domain (Figures 3a and b).
Palmitoylation of hFasL is required for optimal hFasL processing and cytotoxic activity. (a) Diagram of the hFasL transmembrane domain, highlighting the proposed palmitoylation site at aa 82, which we mutated to serine. (b) WSU cells stably transfected with constructs encoding for hFasL, hFasLC82S or hFasLA247E were subjected to an acyl-biotinyl exchange protocol as described in Materials and Methods. Specificity of the experiment was controlled by omitting the hydroxylamine (HA) treatment and Fyn was used as an internal control for reaction efficiency. (c) JH6.2 cells were co-cultured for the indicated time points with WSU cells stably transfected with hFasL, hFasLC82S or mock. Cell death was then quantified by flow cytometric analysis of the subG1 population of propidium iodide-stained ethanol-fixed cells. The graph represents the average of four independent experiments with error bars indicating the S.D. The level of expression of wild-type hFasL or hFasLC82S at the cell surface of WSU transfected cells was comparable (see the inset). (d) Cell lysates of WSU cells stably transfected with constructs encoding for hFasL, hFasLC82S or mock transfected were analyzed by western blotting. (e) Levels of soluble FasL were measured in the supernatant of WSU cells stably transfected with wild-type hFasL or hFasLC82S. The graph represents the average of two independent experiments with error bars indicating the S.D.
To investigate whether binding to the receptor is necessary for hFasL palmitoylation, we tested the palmitoylation status of the hFasLA247E mutant. Our results showed that this mutant, which is unable to bind to Fas, was also palmitoylated (Figure 3b). Therefore, palmitoylation of hFasL in the transmembrane domain at cysteine residue 82 does not require binding to Fas receptor.
We then assessed whether hFasL palmitoylation is important for Fas-mediated cell death. We generated WSU cells stably expressing comparable levels of wild-type hFasL or hFasLC82S at the cell surface (Figure 3c, inset). Co-culture experiments with Fas-expressing JH6.2 target cells revealed that the killing capacity of the hFasLC82S mutant was significantly lower than that of the wild-type hFasL (Figure 3c).
Next, we investigated the proteolysis of the palmitoylation-deficient mutant. Immunoblot analysis of WSU cells stably expressing wild-type hFasL or hFasLC82S revealed that less APL was produced by ADAM10 cleavage of the hFasLC82S mutant compared with wild-type hFasL (Figure 3d). In addition, WSU cells transfected with wild-type hFasL and treated with the general palmitoylation inhibitor 2-bromopalmitate produced decreased amounts of APL compared with untreated cells (Supplementary Figure 1C). Similar to the decreased APL production, significantly lower level of soluble FasL was found in the supernatant of the hFasLC82S mutant cells compared with wild-type hFasL-expressing cells (Figure 3e). Taken together, our data indicate that palmitoylation of hFasL occurs upstream of Fas receptor binding and is a pre-requisite for efficient cleavage by ADAM10.
Neither FasL palmitoylation and nor FasR binding are required for the interaction of hFasL with ADAM10
The ADAM10 pro-form is processed and thereby converted to the mature form of the molecule by the pro-protein convertases furin or PC7. To investigate the nature of the ADAM10 forms interacting with hFasL and the location of this functional interaction, we analyzed the distribution of ADAM10 inside and outside the rafts. Biochemical analysis revealed that the pro-form of ADAM10 was excluded from rafts, whereas the mature form was localized within and outside the raft fractions (Figure 4a). Therefore, confinement of active ADAM10 does not restrict the interaction to any nanodomain while the interaction with the pro-form is restricted to the non-raft fraction.
Palmitoylation of hFasL and receptor-binding are not required for hFasL-ADAM10 interaction. (a) ADAM10 is localized both inside and outside the rafts’ nanodomains. Cells were solubilized in Brij 98 detergent and subjected to sucrose gradient separation before immunoblot analysis of the heavy fraction (HF) and the light fraction (LF) to detect the different forms of hFasL and ADAM10. (b) Co-immunoprecipitation of hFasL with ADAM10. Cell lysates prepared from WSU cells stably transfected with hFasL, hFasLC82S, hFasLA247E or mock were subjected to immunoprecipitation using an anti-ADAM10 antibody and were analyzed by western blot. (c) Stable WSU-hFasL cells were treated with TAPI-2 (50 μM) or Dec-CMK (5 μM, 15 μM) for 4 hours or left untreated (NT) before cell lysates were analyzed by western blot. (d) Levels of sFasL in the supernatant of cells treated with TAPI-2 (50 μM) or Dec-CMK (15 μM) for 4 hours or left untreated (NT) were quantified by ELISA. (e) Stable WSU-hFasL cells were either treated for 4 h with TAPI-2 (50 μM) or Dec-CMK (15 μM), or left untreated (NT) before cell lysates were subjected to immunoprecipitation using an anti-ADAM10 antibody and were analyzed by western blot
To find out whether FasL palmitoylation affects the interaction of hFasL with ADAM10, lysates from WSU cells stably transfected with hFasL, hFasLC82S or hFasLA247E were subjected to immunoprecipitation with an anti-ADAM10 antibody. Figure 4b shows that FasL interacts with the pro-form of ADAM10 regardless of its palmitoylation status. We also observed an increase in ADAM10 binding to hFasLA247E, indicating that this interaction occurs before binding of FasL to its receptor (Figure 4b).
To study the importance of the hFasL interaction with the ADAM10 pro-form in further detail, we treated the cells with the MMP inhibitor TAPI-2 or with the furin inhibitor decanoyl-Arg-Val-Lys-Arg-chlorometylketone (Dec-CMK). Both treatments reduced the amount of cleaved APL fragment and the production of soluble FasL (Figures 4c and d), indicating that the interaction with the ADAM10 pro-form is important for the hFasL cleavage.
We also analyzed the interaction of ADAM10 with hFasL in the presence of these inhibitors. Both treatments led to increased amounts of the FasL co-immunoprecipitated with ADAM10 (Figure 4e). Our results indicate that the interaction with the ADAM10 pro-form is important for the production of APL.
Palmitoylation and receptor interaction alter the nanodomain localization of hFasL
We have previously demonstrated that the localization of hFasL within rafts is essential for maximum induction of cell death.8 In light of these findings, we investigated whether both palmitoylation and receptor binding are required for hFasL raft localization. To monitor hFasL localization of the wild-type FasL and its mutants, we investigated the in vivo lateral diffusion of hFasL-GFP, hFasLA247E-GFP and hFasLC82S-GFP using fluorescence correlation spectroscopy (FCS). FCS measures the apparent diffusion time of molecules, providing information on the diffusion mode of the molecule of interest. On the basis of diffusion law,20 the diffusion time of a protein (τd, the average time a molecule stays within an illuminated area) can be plotted versus the square of the beam radius (w2), and from this representation, the intercept to the y axis can be calculated (t0). The t0 value is indicative of the molecular confinement mode and is positive in the case of confinement to nanodomains, negative in the case of trapping in a meshwork and zero if the molecule is neither confined within discrete domains nor hindered by a network. We have previously reported that wild-type hFasL fused to GFP exhibited a diffusion behavior corresponding to a confinement inside rafts.8 For wild-type FasL, t0 was determined as 35.9±8.5 ms, a diffusion behavior comparable to the raft marker BODIPY-C5-ganglioside-GM1 (25±3 (Lenne et al.21)). In contrast, the palmitoylation-deficient mutant hFasLC82S and the receptor-binding mutant hFasLA247E displayed a significantly different diffusion behavior, reflected by reduced t0 values (8.4±2.9 and 6.2±4.7, respectively), indicating a lower confinement of the proteins into nanodomains (Figure 5).
Wild-type hFasL is confined to nanodomains whereas the palmitoylation- and receptor-deficient mutants are not. Diffusion behavior of hFasL-GFP, hFasLC82S-GFP and hFasLA247E-GFP in COS-7 cells was measured by FCS at 37°C; t0 was determined from the position at which the diffusion curves intersect the time axis (diffusion time). Error bars indicate the S.D. of 10 independent experiments
The effective diffusion coefficient (Deff), defined by the inverse of the diffusion law slope, represents the long-range diffusion of the proteins of interest. Deff was dramatically decreased for the hFasLA247E receptor-binding mutant (0.29±0.02 versus 0.54±0.17 μm2/s for wt hFasL), indicating a greatly reduced long-range diffusion of hFasLA247E.
Discussion
Our results demonstrate that shedding of hFasL by ADAM10 counteracts the cytotoxic activity of hFasL. In this study, we show that ADAM10 processing of hFasL depends on the integrity of the cholesterol- and sphingolipid-enriched nanodomains (rafts) and on the interaction with the Fas receptor. Finally, we have reported the first description of hFasL palmitoylation. This post-translational modification is required for appropriate partitioning of hFasL into rafts, for efficient Fas-mediated cell death and hFasL processing by ADAM10 (Figure 6).
Schematic representation of the ADAM10-mediated processing mechanism of hFasL. At leat two different populations of hFasL were found in our study, one being inside the rafts, the other outside the rafts. Both population can interact with ADAM10, but only the palmitoylated raft localized one might optimally interacts with the receptor and mediates Fas-cell death. Only in this population FasL can be processed; a procedure which downregulates Fas-mediated cell death on the target cell
Data from other groups substantiate our observations that metalloprotease inhibition decreases hFasL processing and increases its killing activity.11 The increased cytotoxicity of hFasL may depend either on the accumulation of the full-length FasL form or on the decrease in the proteolysis products, namely sFasL, APL and SPA. Our results clarify that neither APL nor SPA has an effect on hFasL-induced cell death or on hFasL processing. The decrease in hFasL cytotoxicity is likely due to the reduced amount of full-length FasL and/or the production of soluble FasL. It is possible that sFasL may compete with the full-length form for binding to the Fas receptor. It also remains unclear whether sFasL proteins are exclusively produced by processing of full-length hFasL at the plasma membrane. Overall, the role of hFasL processing by ADAM10 might be a down-modulation or termination of the death signal, as has been described for shedding of receptors, such as TNFR1 following its engagement,22 protection of the neighboring cells by soluble FasL23 or reverse signaling into the ligand-bearing cells, mediated by the liberated SPA fragment.
We demonstrated that FasL interaction with its receptor Fas is necessary for FasL processing. The A247E mutation located within the receptor-binding site, which causes reduced killing of target cells (Supplementary Figure S1A and Del-Rey et al.24), displays reduced FasL processing. hFasL processing following its interaction with Fas would restrict hFasL cytotoxicity to the target cell and avoid excessive cell death. Several ligands and receptors are processed upon binding by their partner, such as the Notch receptor which is cleaved after activation by its ligand Delta (reviewed by Kopan25 and Weinmaster26) and the Ephrin2 ligand, which is processed once it binds to its receptor Eph3A.27 Proteolysis can also be promoted by cross-talk with other receptors. Examples include shedding of the Ephrin ligand which is stimulated by recruitment of G proteins28 and shedding of CD44 which can occur after binding to small hyaluronan oligosaccharides29 or by EGF stimulation through activation of Rac GTPases.30 It remains an open question whether and how other receptors might affect the processing of hFasL. Using the hFasLA247E mutant, which is excluded from rafts, we demonstrated that hFasL recruitment by the Fas receptor is important for its localization into these nanodomains. This hypothesis is further corroborated by our previously published data showing that interaction of hFasL with Fas increases FasL localization inside rafts.8
We then demonstrate that although hFasL and ADAM10 display a similar distribution inside and outside rafts, FasL processing depends on the integrity of the raft domains. Indeed, this result also explains the decreased processing capacity of the receptor-binding mutant hFasLA247E. Individual rafts are relatively small and display heterogeneous protein content. This heterogeneity allows regulation of protein activity owing to differential raft localization.31, 32 In contrast to FasL/ADAM10, differential nanodomain localization of the target protein and the metalloprotease responsible for its cleavage has been reported for other molecules. In these cases, this differential protein localization might control molecule processing. This strategy has been observed for the CD30 protein which is located in rafts, whereas the metalloprotease responsible for its shedding, ADAM17, is excluded.18 In the case of the amyloid precursor protein, the presence of two distinct pools distributed inside and outside of rafts results in differential processing.19
FasL is capable of binding to ADAM10 before interaction with the Fas receptor, and according to our results, this early binding event occurs outside rafts and is important for the proteolysis of hFasL. In this FasL population, hFasL proteolysis is activated by a cleavage of the ADAM10 pro-form by Furin. We cannot exclude the possibility that the FasL cleavage might also occur by direct interaction with the active form of ADAM10. We have yet to determine whether binding to ADAM10 outside rafts is important for FasL processing at the plasma membrane or for intracellular processing. In general, recruitment of ADAM10 to a receptor–ligand system following ligand binding of the receptor EphA3 has also been described. ADAM10 then cleaves the ligand Ephrin 5 in trans.33 Interaction with additional unidentified proteins might also be essential for processing of hFasL by ADAM10 as, for example, recruitment of tetraspanins by ADAM10 has been reported to increase cleavage of ADAM10 substrates34 and/or proteolytic maturation of ADAM10.35
We have previously shown that the death receptors Fas36 and DR437 are palmitoylated. This modification is necessary for receptor partitioning into rafts and for receptor engagement. We demonstrated in this study that FasL palmitoylation also occurs. Indeed, palmitoylation of both ligand and receptor might represent an extended modification for efficient interaction and recruitment. However, palmitoylation of both components is not always necessary, as exemplified by TNF-α, where TNF-α is palmitoylated,38 but its receptor TNFR137 remains unmodified.
Our data imply that palmitoylation of hFasL is important for its biological function, as mutation of the palmitoylation site decreases the level of hFasL-induced cell death. As with the receptor-binding mutant hFasLA247E, the hFasL palmitoylation mutant also exhibits decreased proteolysis by ADAM10. Fluorescence correlation spectroscopy suggests that localization of both the palmitoylation and the receptor-binding mutants is not confined to raft nanodomains, in contrast to the wild-type hFasL. This delocalization would explain the observed decrease in FasL-mediated cell death and proteolytic cleavage.39
In summary, hFasL proteolysis by ADAM10 depends on the interaction of the ligand with its receptor Fas, and the integrity of the raft nanodomains. Post-translational modifications, such as palmitoylation, which change the partitioning of hFasL into rafts, might enable the cell to fine-tune the processing of hFasL by ADAM10 and, consequently, hFasL-mediated cell death.
Materials and Methods
Antibodies and reagents
The antibodies used for immunoblotting were as follows: anti-ADAM10 (Chemicon/Millipore, Billerica, MA, USA), anti-Ezrin (Zymed laboratories, San Francisco, CA, USA), anti-FasL clone G247-4 (BD PharMingen, San Diego, CA, USA, referred to as G247 in the manuscript), anti-FasL clone Ab-3 (Calbiochem/Merck Chemicals, Nottingham, UK, referred to as Ab-3 in the manuscript), anti-FLAG (M2, Sigma), anti-Fyn (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Rab5 (Santa Cruz Biotechnology) and horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies (Jackson ImmunoResearch, West Grove, PA, USA). The anti-FasL antibody clone BR-17 (Diaclone, Besançon, France) was used for the expression analysis by flow cytometry. The matrix metalloprotease inhibitor TAPI-2 (N-(R)-(2-(hydroxyamino-arbonyl)methyl)-4-methylpenthanoyl-L-t-butyl-alanyl-L-alanine, 2-aminoethyl amide) was purchased from Merck (Nottingham, UK). The ADAM10 inhibitor GI254023X ((2R, 3S)-3-(formyl-hydroxyamino)-2-(3-phenyl-1-propyl) butanoic acid ((1S)-2,2-dimethyl-1-methylcar-bamoyl-1-propyl) amide) has been described elsewhere.15 The small molecule protease inhibitor INCB-3619 obtained from P. Scherle (Incyte Corporation, Wilmington, DE, USA) was previously described.16 Furin convertase inhibitor Dec-CMK was purchased from Alexis (Lausen, Switzerland).
Cell culture and transfection procedures
WSU and JH6.2 cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. HEK293 and HEK293 T cells (ATCC no. CRL-1653) were cultured in Dulbecco's Modified Eagle's Medium (Invitrogen), supplemented with 10% FBS at 37°C in a 5% CO2 atmosphere. COS-7 cells (ATCC no. CRL-1657) were grown in DMEM supplemented with 10% FBS, glutamine (2 mM) at 37°C in a 5% CO2 atmosphere.
WSU cells stably expressing hFasL or hFasLC82S were established by transfection with pCR33 hFasL, pCR33 hFasLC82S and pCR33 hFasLA247E constructs using an ECM830 BTX electro square porator (Genotronics, Landgraaf, The Netherlands) at 200 V, 60 ms and 1 pulse, and selected by supplementing the growth medium with 1 mg/ml Geneticin (Gibco/Invitrogen, Carlsbad, CA, USA) 24 h after transfection. HEK293 cells were transiently transfected using calcium phosphate precipitation in 10 cm culture dishes. Transient transfection of COS-7 cells was performed with ExGen 500 (Euromedex, Souffelweyersheim, France).
Constructs
N-terminal FLAG-tagged human FasL was amplified by PCR from a plasmid template and was cloned into the mammalian expression vector pCR33 (kindly provided by Dr. Hermann Eibel, Freiburg, Germany), resulting in pCR33 hFasL. The pCR33 hFasLC82S/pCR33 hFasLA247E constructs were generated using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA), with pCR33 FasL as a template. To produce the N-terminal GFP-tag, FasL cDNA was cloned into the pEGFPC1 vector from Clontech (Mountain View, CA, USA) resulting in pCR33 GFP-FasL. The GFP-hFasLC82S and GFP-hFasLA247E constructs were obtained using the Quikchange site-directed mutagenesis kit, with pCR33 GFP-FasL as a template. Primers utilized are listed in the supplementary materials.
Protein palmitoylation
Protein palmitoylation was determined with an adapted acyl-biotinyl exchange protocol from Drisdel and Green,17 which is comprised of the following three steps. Cell lysates were prepared in 250 mM HEPES, 1 mM EDTA and 2.5% SDS, and incubated with 0.1% methyl methanethiosulfonate (Sigma, St Louis, MO, USA) at 50°C for 20 min to block the free thiols. Proteins were subsequently precipitated with acetone and resuspended in 1 M hydroxylamine pH 7.4 (Sigma) to specifically release thioester-linked palmitoyl moieties and restore the modified cysteines to thiols. The protein suspension was then mixed with 0.2 mM biotin-HPDP (Pierce/Thermo Scientific, Rockford, IL, USA) for 1 h at room temperature. Biotinylated proteins were purified with neutravidin beads (Pierce/Thermo Scientific), separated by SDS-PAGE and submitted to immunoblotting using an antibody-recognizing FasL. Specificity of the experiment was controlled by omitting the hydroxylamine treatment.
Cell death assays
The killing potential of FasL-expressing cells was determined using co-culture assays with Fas-expressing target cells (JH6.2) at a killer-to-target ratio of 1 : 1. Target cells were pre-stained with 5 μM diacetate succinimidyl ester (CFSE, Molecular Probes/Invitrogen, Carlsbad, CA, USA) before co-culturing to distinguish them from killer cells during FACS analysis. The quantification of apoptosis in the target cells was performed by flow cytometry. Briefly, cells were fixed with ice-cold 70% ethanol, washed with 38 mM sodium citrate (pH 7.0) and stained for 20 min at 37°C with 69 μM propidium iodide (Sigma)/38 mM sodium citrate/5 μg per ml RNaseA (Sigma). Cells were subsequently analyzed by flow cytometry (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ, USA). The proportion of apoptotic cells was determined as represented by the subG1 peak.
Separation of detergent-resistant membranes
PNS from 20 × 106 WSU cells was solubilized in 1 ml buffer A (25 mM HEPES, 150 mM NaCl, 1 mM EGTA, 10 mM sodium pyrophosphate, 10 mM sodium floride, 5 mM orthovanadate, protease inhibitor cocktail) containing 1% Brij 98 (Aldrich, St Louis, MO, USA) and 10% glycerol for 1 h on ice, followed by the addition of 2 ml of 2 M sucrose in buffer A, before being placed at the bottom of a step sucrose gradient (1.33-0.9-0.8-0.75-0.7-0.6-0.5-0.4-0.2 M) in buffer A. The gradient was centrifuged at 250 000 × g for 16 h in an SW41 Beckman rotor (Beckman Instruments, Gagny, France) at 4°C. One-milliliter fractions were harvested from the top, except for the last fraction that contained 3 ml.
Fluorescence correlation spectroscopy experiments
Fluorescence correlation spectroscopy (FCS) measurements were performed with a custom apparatus as previously described. Briefly, FCS data were collected using a confocal-based microscope (Axiovert 200M; Zeiss, Oberhocken, Germany) equipped with an Aprochromat 40 × /1.2 numeric aperture water-immersion objective and excitation from a 488-nm line of an Ar+ ion laser. The laser waist was set by selecting the lateral extension of laser beam falling into the back aperture of the objective with a diaphragm. Fluorescence was collected through a 525 to 565-nm bandpass filter. A confocal pinhole reduced the out-of-focus fluorescence. FCS measurements were carried out in Hanks balanced salt solution with 10 mM HEPES, pH 7.4, by illuminating the sample with an excitation power of 3 μW at the back aperture of the objective lens. Autocorrelations were processed by a hardware correlator (http://www.correlator.com, Hong Kong) and data were analyzed with built-in functions of IGOR Pro (WareMetrics, Lake Oswego, OR, USA).
Immunoprecipitation
WSU cells (20 × 106) were sonicated in buffer A (25 mM HEPES, 150 mM NaCl, 1 mM EGTA, 10 mM NaP-P, 10 mM NaF, 5 mM Na3VO4, 25 mM β-glycerol phosphate, 5 μg/ml leupeptin, 0.125U α2 macroglobulin, 10 μg/ml pepstatin A, 10 μg/ml chymostatin) containing 0.5% Nonidet P-40 (NP-40) and 10% glycerol at 43°C and then centrifuged to discard cell debris. The supernatant was incubated overnight at 4°C with 30 μl Protein G sepharose beads (Invitrogen) pre-incubated with an anti-ADAM10 antibody (Millipore). The beads were washed four times with buffer A containing 0.5% NP-40 and 10% glycerol and the immunoprecipitates were eluted from the beads with Laemmli buffer at 95°C for 5 min. Samples were separated by SDS-PAGE and analyzed by immunoblotting.
FasL-specific ELISA
The culture medium of WSU cells incubated at 10 × 106 cells per ml for 4 h was harvested to analyze soluble proteins. sFasL present in the supernatant was measured by ELISA using the human sFasL module set (Bender Medsystems, Vienna, Austria). Briefly, a 96-microwell plate was coated overnight with 2.5 μg/ml mouse anti-FasL capture antibody at 4°C, washed with phosphate-buffered saline (PBS)/0.05% Tween (PBS-T) and then blocked with 3% bovine serum albumin in PBS for 2 h at room temperature. After washing with PBS-T, samples and standard were added. Next, biotinylated anti-FasL detection antibody was applied to each well and the plate was incubated for 2 h at room temperature, washed and incubated for 1 h at room temperature with a streptavidin-peroxidase conjugate. The TMB super-sensitive microwell one component peroxidase substrate (Tebu-bio, Le-Perray-en-Yvelines, France) was added and the reaction was stopped after 20 min incubation at room temperature by the addition of 4 N H2SO4 and the microwell plate was read at 450 nm.
Abbreviations
- ADAM10:
-
A disintegrin and metallopeptidase 10
- APL:
-
ADAM10-processed FasL
- EGF:
-
epidermal growth factor
- FasL:
-
Fas ligand
- FCS:
-
fluorescence correlation spectroscopy
- GFP:
-
green fluorescence protein
- MMP:
-
matrix metalloproteinase
- NK:
-
natural killer
- PNS:
-
post-nuclear supernatant
- sFasL:
-
soluble Fas ligand
- SPA:
-
SPPL2a-processed APL
- SPPL2a:
-
signal peptide peptidase-like 2A
- TNF:
-
tumor necrosis factor
References
Janssen O, Qian J, Linkermann A, Kabelitz D . CD95 ligand—death factor and costimulatory molecule? Cell Death Differ 2003; 10: 1215–1225.
Alderson MR, Tough TW, Davis-Smith T, Braddy S, Falk B, Schooley KA et al. Fas ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med 1995; 181: 71–77.
Griffith TS, Brunner T, Fletcher SM, Green DR, Ferguson TA . Fas ligand-induced apoptosis as a mechanism of immune privilege. Science 1995; 270: 1189–1192.
Ferguson TA, Green DR, Griffith TS . Cell death and immune privilege. Int Rev Immunol 2002; 21: 153–172.
Krammer PH . The tumor strikes back: new data on expression of the CD95(APO-1/Fas) receptor/ligand system may cause paradigm changes in our view on drug treatment and tumor immunology. Cell Death Differ 1997; 4: 362–364.
Restifo NP . Not so Fas: re-evaluating the mechanisms of immune privilege and tumor escape. Nat Med 2000; 6: 493–495.
Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med 2002; 195: 1303–1316.
Cahuzac N, Baum W, Kirkin V, Conchonaud F, Wawrezinieck L, Marguet D et al. Fas ligand is localized to membrane rafts, where it displays increased cell death-inducing activity. Blood 2006; 107: 2384–2391.
Nachbur U, Kassahn D, Yousefi S, Legler DF, Brunner T . Posttranscriptional regulation of Fas (CD95) ligand killing activity by lipid rafts. Blood 2006; 107: 2790–2796.
Kirkin V, Cahuzac N, Guardiola-Serrano F, Huault S, Luckerath K, Friedmann E et al. The Fas ligand intracellular domain is released by ADAM10 and SPPL2a cleavage in T-cells. Cell Death Differ 2007; 14: 1678–1687.
Schulte M, Reiss K, Lettau M, Maretzky T, Ludwig A, Hartmann D et al. ADAM10 regulates FasL cell surface expression and modulates FasL-induced cytotoxicity and activation-induced cell death. Cell Death Differ 2007; 14: 1040–1049.
Tanaka M, Itai T, Adachi M, Nagata S . Downregulation of Fas ligand by shedding. Nat Med 1998; 4: 31–36.
Knox PG, Milner AE, Green NK, Eliopoulos AG, Young LS . Inhibition of metalloproteinase cleavage enhances the cytotoxicity of Fas ligand. J Immunol 2003; 170: 677–685.
O’Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature 2009; 461: 659–663.
Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 2003; 102: 1186–1195.
Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 2006; 10: 39–50.
Drisdel RC, Green WN . Labeling and quantifying sites of protein palmitoylation. Biotechniques 2004; 36: 276–285.
von Tresckow B, Kallen KJ, von Strandmann EP, Borchmann P, Lange H, Engert A et al. Depletion of cellular cholesterol and lipid rafts increases shedding of CD30. J Immunol 2004; 172: 4324–4331.
Ehehalt R, Keller P, Haass C, Thiele C, Simons K . Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol 2003; 160: 113–123.
Wawrezinieck L, Rigneault H, Marguet D, Lenne PF . Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J 2005; 89: 4029–4042.
Lenne PF, Wawrezinieck L, Conchonaud F, Wurtz O, Boned A, Guo XJ et al. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J 2006; 25: 3245–3256.
McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 1999; 97: 133–144.
Hallermalm K, De Geer A, Kiessling R, Levitsky V, Levitskaya J . Autocrine secretion of Fas ligand shields tumor cells from Fas-mediated killing by cytotoxic lymphocytes. Cancer Res 2004; 64: 6775–6782.
Del-Rey M, Ruiz-Contreras J, Bosque A, Calleja S, Gomez-Rial J, Roldan E et al. A homozygous Fas ligand gene mutation in a patient causes a new type of autoimmune lymphoproliferative syndrome. Blood 2006; 108: 1306–1312.
Kopan R . Notch: a membrane-bound transcription factor. J Cell Sci 2002; 115: 1095–1097.
Weinmaster G . Notch signal transduction: a real rip and more. Curr Opin Genet Dev 2000; 10: 363–369.
Hattori M, Osterfield M, Flanagan JG . Regulated cleavage of a contact-mediated axon repellent. Science 2000; 289: 1360–1365.
Ohtsu H, Dempsey PJ, Eguchi S . ADAMs as mediators of EGF receptor transactivation by G protein-coupled receptors. Am J Physiol Cell Physiol 2006; 291: C1–10.
Sugahara KN, Murai T, Nishinakamura H, Kawashima H, Saya H, Miyasaka M . Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J Biol Chem 2003; 278: 32259–32265.
Murai T, Miyauchi T, Yanagida T, Sako Y . Epidermal growth factor-regulated activation of Rac GTPase enhances CD44 cleavage by metalloproteinase disintegrin ADAM10. Biochem J 2006; 395: 65–71.
Pralle A, Keller P, Florin EL, Simons K, Horber JK . Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol 2006; 148: 997–1008.
Coskun U, Simons K . Membrane rafting: from apical sorting to phase segregation. FEBS Lett 2010; 584: 1685–1693.
Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E et al. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 2005; 123: 291–304.
Arduise C, Abache T, Li L, Billard M, Chabanon A, Ludwig A et al. Tetraspanins regulate ADAM10-mediated cleavage of TNF-alpha and epidermal growth factor. J Immunol 2008; 181: 7002–7013.
Xu D, Sharma C, Hemler ME . Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. FASEB J 2009; 23: 3674–3681.
Chakrabandhu K, Herincs Z, Huault S, Dost B, Peng L, Conchonaud F et al. Palmitoylation is required for efficient Fas cell death signaling. EMBO J 2007; 26: 209–220.
Rossin A, Derouet M, Abdel-Sater F, Hueber AO . Palmitoylation of the TRAIL receptor DR4 confers an efficient TRAIL-induced cell death signalling. Biochem J 2009; 419: 185–192.
Utsumi T, Takeshige T, Tanaka K, Takami K, Kira Y, Klostergaard J et al. Transmembrane TNF (pro-TNF) is palmitoylated. FEBS Lett 2001; 500: 1–6.
Brown DA . Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 2006; 21: 430–439.
Acknowledgements
We thank K Chakrabandhu, H Peterfy and E Doma for advices and helpful discussions. This work was supported by institutional funds from the Centre National de la Recherche Scientifique (CNRS), and by grants from the Ligue nationale contre le cancer (LNCC), the Association pour la Recherche contre le Cancer (ARC), the Institut National du Cancer (INCa), the Agence Nationale de la Recherche (ANR-08-PCVI-0034-01). FGS was supported by ANR, and AR by a fellowship from the Ligue nationale contre le cancer (LNCC), MZ by grants from the German Research Foundation (ZO110/4-1, ZO110/2-1/2) and KL by a grant from the German National Academic Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies the paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Guardiola-Serrano, F., Rossin, A., Cahuzac, N. et al. Palmitoylation of human FasL modulates its cell death-inducing function. Cell Death Dis 1, e88 (2010). https://doi.org/10.1038/cddis.2010.62
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2010.62
Keywords
This article is cited by
-
ADAM10-a “multitasker” in sepsis: focus on its posttranslational target
Inflammation Research (2023)
-
Palmitoylation is required for TNF-R1 signaling
Cell Communication and Signaling (2019)
-
Impairment of Fas-ligand–caveolin-1 interaction inhibits Fas-ligand translocation to rafts and Fas-ligand-induced cell death
Cell Death & Disease (2018)
-
Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability
Cell Death & Differentiation (2015)
-
Analysis of apoptosis methods recently used in Cancer Research and Cell Death & Disease publications
Cell Death & Disease (2012)