Abstract
Sphingolipids are involved in several cellular functions, including maintenance of cell wall integrity. To gain insight into the role of individual genes of sphingolipid biosynthetic pathway, we have screened Saccharomyces cerevisiae strains deleted in these genes for sensitivity to cell wall perturbing agents calcofluor white and congo red. Only deletants of FEN1 and SUR4 genes were found to be sensitive to both these agents. Candida albicans strains deleted in their orthologs, CaFEN1 and CaFEN12, respectively, also showed comparable phenotypes, and a strain deleted for both these genes was extremely sensitive to cell wall perturbing agents. Deletion of these genes was reported earlier to sensitise cells to amphotericin B (AmB), which is a polyene drug that kills the cells mainly by binding and sequestering ergosterol from the plasma membrane. Here we show that their AmB sensitivity is likely due to their cell wall defect. Further, we show that double deletant of C. albicans is defective in hyphae formation as well as biofilm development. Together this study reveals that deletion of FEN1 and SUR4 orthologs of C. albicans leads to impaired cell wall integrity and biofilm formation, which in turn sensitise cells to AmB.
Similar content being viewed by others
Introduction
Candida albicans, an opportunistic pathogen of humans, can cause infections ranging from superficial skin infections to life-threatening invasive infections1,2. Mortality due to invasive infections can be as high as 75% worldwide3,4. Though several antifungals are available for treatment, they suffer from lack of broad spectrum of activity, drug resistance or high cost4. Since fungi are eukaryotes they share a large number of cell processes with their mammalian hosts and thus the number of drug targets are limited5. However, ergosterol and cell wall that are present in fungi but absent in their hosts serve as targets of commonly used antifungals5,6. While antifungals such as azoles and terbinafine inhibit ergosterol biosynthesis, amphotericin B, a polyene antifungal, binds to ergosterol of plasma membrane and kills the cells primarily by sequestering ergosterol5,7,8. Among antifungals that target cell wall, echinocandins and nikkomycin inhibit β-Glucan synthesis and chitin synthesis, respectively. Defective cell wall biogenesis is known to attenuate infections by C. albicans9,10. Thus, a better understanding of cell wall biogenesis and integrity may reveal novel targets for antifungal drug development.
Yeast cell wall is a complex network of polysaccharides (β-1,3-glucan, β-1,6-glucan and chitin) and mannoproteins11,12. β-1,3-glucan and mannoproteins are major components of cell wall comprising 30–45% and 30–50%, respectively. β-1,6-glucan and chitin contribute to 5–10% and 1.5–6.0% of the total cell wall biomass11. The cell wall is involved in several protective functions of the cells, including stabilisation of intracellular osmotic balance, oxidative stress, heat stress and antifungal resistance11,12,13,14,15. Cell wall integrity (CWI) is maintained by PKC1–MPK1 (Slt2) pathway, which helps in the biogenesis of cell wall16,17,18,19. Sphingolipid pathway intermediates, such as dihydrosphingosine and phytosphingosine, are also involved in CWI signalling20,21. Sphingolipids are a class of sphingoid backbone or long chain base (LCB) containing lipids21,22. These are major components of eukaryotic membranes and are abundant in the plasma membrane. In S. cerevisiae, these constitute 30% of the total phospholipids and about 7% of the total mass of the plasma membrane23. Sphingolipids coexist with sterols and glycerophospholipids and define the integrity as well as the plasticity of plasma membranes24,25. In addition to providing a structural framework, along with sterols they form functional microdomains in plasma membrane called lipid rafts26,27, which are involved in many physiological activities of the cells, including actin organisation, endocytosis, signal transduction and cell morphogenesis28,29,30,31. Moreover, lipid rafts participate in the sorting of glycosylphosphatidylinositol (GPI)-anchor proteins to the cell surface32,33,34. About one-third of the identified GPI-anchor proteins of S. cerevisiae contributes to cell wall biogenesis and their deficiency results in cell wall defect12.
Though previous studies have shown the involvement of sphingolipids in the CWI signalling20,21, there are no reports regarding the role of individual sphingolipid biosynthetic pathway genes in CWI modulation. To identify these genes, first, we have screened deletion mutants of sphingolipid biosynthetic pathway genes of S. cerevisiae with cell wall perturbing agents calcofluor white (CFW) and congo red (CR) and found that only deletants of FEN1 and SUR4 genes were sensitive to both the chemicals. These strains also showed other phenotypes typical of mutants with impaired CWI. Next, we have checked if the orthologs of these genes in C. albicans have a similar role, and found that their mutants also have comparable phenotypes. Moreover, C. albicans strain deleted in both these genes was found to be defective in hyphae formation and biofilm development. Since these mutants are also amphotericin B (AmB) sensitive35, we have tested the correlation between these phenotypes and find that their AmB sensitivity is likely due to their impaired cell wall.
Results and Discussion
Deletants of FEN1 and SUR4 genes of S. cerevisiae and their orthologs in C. albicans are impaired in cell wall integrity
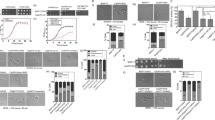
Though the role of sphingolipids in CWI signalling is known20,21, that of individual sphingolipid biosynthetic pathway genes is not yet reported. To identify such genes, we have screened homozygous deletants of twenty-two non-essential genes of the sphingolipid pathway of S. cerevisiae (Fig. S1), which were constructed as part of the yeast deletion project36, at various concentrations of CFW and CR. Cells with defective cell wall are known to increase chitin synthesis as a compensatory mechanism to maintain the cell wall integrity11,12. CFW and CR, which bind chitin, have been extensively used to identify such mutants since they are sensitive to a lower concentration of these compounds compared to normal cells37,38. Two deletants, fen1Δ/Δ and sur4Δ/Δ, were found to be sensitive to both the chemicals compared to the parent strain (BY4743) (Fig. 1). CFW sensitivity of FEN1 deletant was also reported earlier, after being identified through a screen for genetic interaction with CCW12, a gene involved in CWI39. Among other deletants, lcb3Δ/Δ and sur2Δ/Δ were found to be slightly sensitive to CFW (Fig. 1a), but their growth on CR was comparable to the parent strain (Fig. 1b). While we cannot rule out moderate cell wall defect in these mutants, we initially focused our study on FEN1 and SUR4 deletants, since only these were sensitive to both CFW and CR, and thus definitely impaired in cell wall integrity. Heterozygous deletants of essential genes of sphingolipid biosynthetic pathway were also screened with the notion that if they are haploinsufficient, then their CFW and CR sensitivity can be determined. However, their growth was comparable to the parent strain (Fig. 1). FEN1 (ELO2) and SUR4 (ELO3) along with ELO1 encode fatty acid elongases, which synthesise long chain or very long-chain fatty acids (LCFA or VLCFA)21,40,41. Elo1p, Elo2p (Fen1p) and Elo3p (Sur4p) are involved in the synthesis of C14 to C16 LCFA, up to C24 VLCFA, and C24 or C26 VLCFA respectively21,40,41,42 and mutations in FEN1 and SUR4 genes result in shortened fatty acid chains and lower levels of sphingolipids40,42.
An overview of sphingolipid biosynthetic pathway is shown in Fig. S1. Ten-fold serial dilutions of cells were spotted onto synthetic complete agar plates with indicated concentration of (a) calcofluor white (CFW), or (b) congo red (CR). Plates were incubated at 30 °C for 2 days before being photographed.
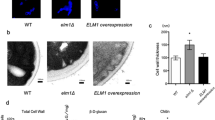
Since cell wall mutants that are sensitive to CFW and CR often have more chitin in their cell wall11,12,37, we used CFW staining to visualise chitin. More intense and larger area of fluorescence was seen at bud scars and mother-daughter cell junctions of Scfen1Δ/Δ and Scsur4Δ/Δ strains. Semi-quantification of fluorescence intensity using the NIS-Elements software showed that Scfen1Δ/Δ and Scsur4Δ/Δ strains have 27% and 32%, respectively, higher intensity than the parent strain, confirming that these mutants indeed have more chitin (Fig. 2a). Since chitin level is also increased in mutants impaired in the synthesis of β-1,3-glucan12,43, we speculated that Scfen1Δ/Δ and Scsur4Δ/Δ strains may be defective in β-1,3-glucan synthesis. Cells with decreased levels of ß-1,3-glucan, or increased accessibility of β-1,3-glucan due to some other defect in the cell wall, are more sensitive to zymolyase having β-1,3-glucanase as its principal constituent44. Scsur4Δ/Δ strain was found to be sensitive to zymolyase compared to the parent strain and Scfen1Δ/Δ strain was even more sensitive (Fig. 2b), which is comparable to their relative sensitivity to CFW and CR (Fig. 1). Next, we tested the sensitivity of these deletants to SDS, a detergent usually employed to determine the compactness of the cell wall, since less compact cell wall would allow SDS to readily reach and damage the plasma membrane resulting in cell death9,45. Scsur4Δ/Δ strain was found to be SDS-sensitive (Fig. 2c) suggesting that it has a less compact cell wall structure; however, altered lipid composition of the plasma membrane might have also rendered it more susceptible to SDS.
(a) Deletion of FEN1 and SUR4 genes increases chitin content in the cell wall. Cells of the parent (BY4743) and deletants were stained with CFW, and images were captured at identical conditions with a fluorescence microscope using 100× objective lens (upper panel). DIC images of corresponding fields are shown in the lower panel. (b) fen1Δ/Δ and sur4Δ/Δ strains are more sensitive to zymolyase. Zymolyase digestion of the parent and deletants was monitored by periodically measuring OD600nm until 90 minutes. Average values of two independent experiments, carried out in triplicate each time, are shown. (c) SDS sensitivity of fen1Δ/Δ and sur4Δ/Δ strains. Cells were spotted onto synthetic complete agar plates with indicated concentration of SDS and incubated at 30 °C for 2 days before being photographed.
CaFEN1 and CaFEN12 are orthologs of ScFEN1 and ScSUR4, respectively, in C. albicans35. To check if these are also involved in cell wall integrity, we tested the sensitivity of their deletion mutants to cell wall perturbing agents. Similar to the deletants of ScFEN1 and ScSUR4, deletants of CaFEN1 and CaFEN12 showed comparable sensitivity to CFW. Moreover, the strain deleted in both CaFEN1 and CaFEN12 was found to be hypersensitive to CFW (Fig. 3a). To check that the phenotypes seen with the double delete strain are actually due to deletion of these genes, and not because of any extraneous mutation, reintegrant strains were constructed by introducing wild-type CaFEN1 or CaFEN12 genes at their respective loci in the double delete strain. This has resulted in the suppression of the sensitivity of the double delete strain to AmB, CFW and CR (Fig. S2), confirming that the phenotypes of this strain are in fact due to deletion of these two genes. The double delete strain also showed very intense fluorescence after CFW staining (Fig. 3b); by semi-quantification, the fluorescence intensity of this strain was found to be 127% higher than that of the wild-type strain SN95. The Cafen1Δ/Δ and Cafen12Δ/Δ strains, respectively, showed 46% and 17% increase in fluorescence. These results indicate higher chitin content in the cell wall, particularly in the double delete strain. We also tested the sensitivity of these deletants to zymolyase, and as expected double deletant was more sensitive compared to single deletants and parent strain (Fig. 3c), and their relative sensitivity was comparable to their sensitivity to CFW (Fig. 3a), suggesting that chitin deposition is associated with defective synthesis or increased accessibility of β-1,3-glucan. Compactness of the cell wall was also assessed in these deletants by SDS sensitivity test. Double deletant was not growing at the tested concentrations of SDS, and similar to Scsur4Δ/Δ deletant, Cafen12Δ/Δ was more sensitive to SDS compared to Cafen1Δ/Δ and the parent strain (Fig. 3d). Altogether, these results reveal the functional importance of Fen1p and Sur4p in maintaining the CWI in both S. cerevisiae and C. albicans. Though double deletion of FEN1 and SUR4 orthologs is not lethal in C. albicans, unlike in S. cerevisiae46, synergistic sensitivity towards cell wall perturbing agents by deletion of CaFEN1 and CaFEN12 in C. albicans suggest that both these genes independently impair CWI. Individual deletion of FEN1 and SUR4 genes of C. glabrata and S. cerevisiae differentially affect susceptibility to echinocandins caspofungin and micafungin47. Moreover, the double delete (Cafen1Δ/Δ Cafen12Δ/Δ) strain of C. albicans also showed a similar phenotype48. Since echinocandins inhibit the 1,3-β glucan synthase activity, changes in the efficacy of these antifungals against these mutants further confirm the role of these genes in CWI.
(a) C. albicans strains deleted in CaFEN1 and CaFEN12 genes are hypersensitive to CFW. Cells of the parent (SN95) and single as well as double deletant strains of CaFEN1 and CaFEN12 genes were spotted onto synthetic complete agar plates with indicated concentration CFW and incubated at 30 °C for 2 days. (b) Chitin level of single and double deletants of CaFEN1 and CaFEN12 genes. After CFW staining images were captured under identical conditions with a fluorescence microscope using 100× objective lens. (c) Zymolyase sensitivity, determined as described in Fig. 2b. (d) SDS sensitivity. Cells of the parent and deletants were incubated with indicated concentration of SDS as mentioned in Fig. 2c.
Compromised sphingolipid biosynthesis leads to cell wall defect
To gain further insight into the functional significance of sphingolipids for CWI, we checked the cell wall defect in the presence of myriocin, which is an inhibitor of serine palmitoyltransferase that catalyses the first committed step of sphingolipid biosynthesis21,22. Wild-type strains of S. cerevisiae (FY4, BY4743 and BY4741) and C. albicans (SC5314) were used to test myriocin effect on CWI. Myriocin, at sub-lethal concentration (0.2 μg/ml), rendered the cells sensitive to CFW (100 μg/ml), indicating cell wall defect (Fig. 4). To check if the cell wall defect was because of depletion of sphingolipids, we supplemented phytosphingosine (PHS; 10 μM) to combined myriocin and CFW treated cells. PHS is a downstream intermediate in sphingolipid pathway and is known to rescue the myriocin mediated inhibition of sphingolipid biosynthesis49. PHS reversed the myriocin mediated CFW sensitivity (Fig. 4). We have also tested the effect of myriocin and PHS on CFW sensitivity of delete strains (Fig. S3). While the deletants are as such more sensitive to CFW compared to wild-type strains (second panel), addition of myriocin further sensitised them to CFW (fourth panel). PHS seems to reverse only the additional sensitivity caused by myriocin (fifth panel), suggesting that it does not compensate for the lack of elongase activities of Fen1 and Sur4, though it suppresses depletion of sphingolipids and CFW sensitivity caused by myriocin (Fig. 4). These results confirm that the cell wall defect was indeed due to the depletion of sphingolipids.
Wild-type strains of S. cerevisiae (BY4741, BY4743, FY4) and C. albicans (SC5314) were tested for their sensitivity to CFW alone or in the presence of myriocin or myriocin and phytosphingosine (PHS). The sublethal concentration of myriocin sensitised the cells to CFW, which is rescued by supplementation with PHS.
Cell wall defect is likely responsible for amphotericin B sensitivity of FEN1 and SUR4 deletants
We have previously reported that deletion of FEN1 and SUR4 genes leads to AmB sensitivity in S. cerevisiae, which was further validated in C. albicans deleted for their orthologs CaFEN1 and CaFEN12, respectively35. Since alterations in the cell wall composition are known to affect AmB susceptibility50,51,52, it is likely that AmB sensitivity of these deletants is due to their cell wall defect. To check if directly modulating cell wall integrity would affect AmB sensitivity, we have tested the effect of CFW on AmB sensitivity. CFW was found to sensitise cells to AmB (Fig. 5), indicating cell wall damage can result in AmB sensitivity. To further assess the contribution of cell wall damage to AmB sensitivity, we tested deletants of three other genes that are not part of sphingolipid biosynthetic pathway but involved in CWI, for AmB sensitivity phenotype. FKS1, GAS1 and KRE6 are critical for cell wall biogenesis encoding β-1,3-glucan synthase, 1,3-beta-glucanosyltransferase, and type II integral membrane protein required for beta-1,6 glucan biosynthesis, respectively53. β-1,3-glucan synthase synthesises the β-1,3-glucan, which is further elongated and arranged in the side chains by 1,3-beta-glucanosyltransferase activity. KRE6 encoded protein participates in the synthesis of β-1,6-glucan, which cross-links to the side chains of β-1,3-glucan and provides tight mesh structures of the cell wall. Deletion of these genes is known to be associated with cell wall defect and loss of CWI53, but their AmB sensitivity phenotype has not been reported so far. We tested AmB susceptibility of fks1Δ/Δ, gas1Δ/Δ and kre6Δ/Δ strains of S. cerevisiae and found that these were, in fact, sensitive to AmB (Fig. S4a). Normal transport of Gas1p and the other GPI-anchored proteins from ER to Golgi is reported to be abrogated in the deletants of FEN1 and SUR4 genes because of reduced sphingolipid biosynthesis54. Thus it appears likely that the AmB sensitivity of FEN1 and SUR4 mutants is due to weakened cell wall arising out of defective Gas1p transport. However, as these mutants would have shortened fatty acid chains and lower levels of sphingolipids40,42, we cannot rule out the contribution of these changes in the membrane lipid composition to the AmB sensitivity of cells. Overexpression of PMP3 gene, encoding Plasma Membrane Proteolipid 3 protein, enhances AmB resistance55,56, which is also dependent on sphingolipid biosynthetic pathway55. Deletion of this gene renders the cells hypersensitive to AmB55,56, but unlike in FEN1 or SUR4 deletants, this does not involve any change in CWI55.
Several yeast genes such as PKC1, which are involved in CWI, when mutated render the cells osmotically fragile19. To check whether osmotic imbalance contributes to AmB sensitivity of fks1Δ/Δ, gas1Δ/Δ and kre6Δ/Δ strains, we tested them in the presence of sorbitol as osmotic support. Sorbitol failed to rescue the AmB sensitivity (Fig. S4a), implying that this phenotype is not due to osmotic imbalance. We also tested AmB sensitivity of FEN1 and SUR4 deletants of S. cerevisiae in the presence of sorbitol and found that their sensitivity was also not rescued by sorbitol (Fig. S4b). However, AmB sensitivity of C. albicans single gene deletants was partially suppressed by sorbitol (Fig. S4b), indicating that deletion of CaFEN1 and CaFEN12 leads to cell wall defect accompanied with osmotic destabilisation.
Candida albicans strain deleted in both CaFEN1 and CaFEN12 is defective in hyphae and biofilm formation
Deletion of both FEN1 and SUR4 genes in S. cerevisiae is lethal. However, both their orthologs could be deleted in C. albicans without loss of viability, but the strain was slow growing35. In a further phenotypic analysis of these strains, we tested their hyphal growth in solid medium (YPD agar) under hypha-inducing condition (10% FBS). While double deletant was deficient in hypha formation and invasive growth, the phenotypes of single deletants were comparable to the parent strain (Fig. 6a and b). Hyphal growth is a characteristic feature of biofilm development, and both are important for pathogenicity of C. albicans57,58,59. Biofilm is a complex three-dimensional structure consisting of yeast, pseudohyphae and hyphae, which are usually surrounded by a protective layer called extracellular matrix substance that adheres to the surfaces57,60. Immunocompromised patients with indwelling medical devices are more prone to Candida biofilm development57,61,62. Biofilm formation is considered to be one of the leading causes of candidiasis mediated mortality because of their natural antifungal resistance57,61,62,63. Since hyphae are integral to biofilm architecture, and hyphae formation is impaired in Cafen1Δ/Δ Cafen12Δ/Δ double deletant, we tested the effects of single as well as double deletion of CaFEN1 and CaFEN12 on biofilm formation in a microtiter plate with colorimetric XTT reduction assay. Biofilm formation was reduced by 70% in the double deletant as compared to the parent strain though no effect was seen for single deletants (Fig. 6c d and). Formed biofilms were also visualised by scanning electron micrography. While parent and single deletant strains showed typical biofilm architecture with extensive hyphae, double deletant strain lacked hyphal growth or biofilm formation but formed pseudohyphal like structure (Fig. 7). Inhibition of sphingolipid biosynthesis was reported to affect lipid rafts, hyphae and biofilm formation64. Thus, the defect in hyphal growth and biofilm formation in double deletant is likely due to impaired sphingolipid biosynthesis.
(a) Hyphae formation assay on YPD agar plate containing 10% fetal bovine serum. (b) Invasive growth into agar. After washing the cells on the agar surface with water, the agar was vertically sliced and observed under a microscope. (c) Biofilm formation. For each strain cells at a density of 1 × 106 cells/ml were dispensed into 96-well microtiter plate in quadruplicate and incubated at 37 °C for 2 days. Then nonadherent cells were removed by washing, and leaving behind mature biofilm in the wells (upper panel). Metabolic activity of biofilms was visualised by conversion of XTT from light orange to dark orange colour (lower panel). (d) Quantification of biofilm formation by colorimetric XTT reduction assay. Average values of XTT reduction reading at 492 nm of each strain is expressed as a percentage of the value of parent strain. Error bars represent means ± standard deviations of three independent quadruplicate experiments.
Previously35, as well as in this study, we showed that the planktonic cells of these deletants were more sensitive to the AmB as compared to the parent strain. Since biofilms are inherently more resistant to antifungals57,61,62,63, we tested AmB sensitivity of these deletants in preformed biofilms and during biofilm formation. We incubated preformed biofilms of the deletants and parent strain with AmB at 37 °C for 2 days and then determined the viability by XTT reduction assay. The AmB sensitivity of biofilms formed by single deletants was comparable to the parent strain. However, double deletant was 8-fold more sensitive to AmB compared to parent strain suggesting that the defect in biofilm formation rendered the cells more sensitive to AmB (Table 1). Moreover, we also tested the AmB sensitivity during biofilm formation, in which cell density comparable to that of biofilm formation was used. AmB was included from the beginning and incubation was done at 37 °C for 2 days to allow the formation of mature biofilms. At the end of incubation, non-adherent cells were aspirated and washed out, and remaining formed biofilms were quantified by XTT assay. During biofilm formation, single and double deletants were found to be 2-fold and 8-fold, respectively, more sensitive to AmB compared to the parent strain (Table 1). The enhanced sensitivity of double deletant appears to be due to its inability to form biofilms that are inherently more resistant to AmB57,61,62,63. Biofilm extracellular matrix constituents such as β-1,3-glucan and extracellular DNA are known contributing factors for AmB resistance62.
In conclusion, we have shown that C. albicans genes CaFEN1 and CaFEN12 involved in sphingolipid biosynthesis are critical for cell wall integrity and for the formation of hyphae and biofilm. The strain deleted in both these genes is highly sensitive to AmB, likely due to its weak cell wall and inability to form biofilm.
Materials and Methods
Yeast strains, media and growth conditions
Strains of S. cerevisiae and C. albicans used in this study are listed in Table 2. YPD, Synthetic complete (SC) and RPMI-1640 media were prepared and used as described previously35. For primary (overnight) or exponential culture strains were grown in YPD broth at 30 °C with agitation (200 rpm). Stock solutions of CFW (sigma), CR (sigma), AmB (Sigma), myriocin (Sigma) and PHS (TCI chemicals) were prepared in dimethyl sulfoxide (Sigma) and stored at −20 °C until use.
Generation of CaFEN1 and CaFEN12 reintegration strains
Wild-type copy of CaFEN1 was amplified from C. albicans SC5314 genomic DNA using primers CaFEN1-US1 (5′-CAATCATCGCACATAAAACC) and CaFEN1-DA2 (5′-GGTGATACATTTTTCGGAG). Similarly, wild-type copy of CaFEN12 was amplified from the genomic DNA using primers CaFEN12-US1 (5′-ATAATGGAAGAGGGAAGGC) and CaFEN12-DA2 (5′- GTCATGTAGTTCCTGCTACC). Both the PCR products (0.5–1 μg) were separately transformed into the double delete strain SN95F1F12. The transformants were selected at 42 °C, a temperature at which the reintegrant strains would grow, but not the double delete strain48. The integration of CaFEN1 and CaFEN12 at their target loci was confirmed by diagnostic PCR using primers CaFEN1-DG-S (5′-CTCAATAGTCATCGACACG) and CaFEN1-DG-R1 (5′-GTGGTAGTCAAACCACTCCAC) for CaFEN1 and primers CaFEN12-DG-S (5′-GAAGGATATGGAACATTCG) and CaFEN12-DG-R1 (5′-TCCATACTGCTCATGTTGAAG) for CaFEN12.
Susceptibility testing by dilution spotting
Overnight grown yeast strains in YPD broth were re-inoculated in fresh YPD medium and incubated at 30 °C with shaking at 200 rpm. The exponential cells were harvested, washed with water and normalised to an optical density (OD600nm) of 1.0 (2 × 107 cells/ml). These cell suspensions were ten-fold serially diluted, and 5 μl of each dilution was spotted on SC agar plates containing different concentrations of tested chemicals or drugs. Growth was assessed by incubating the plates at 30 °C for 2 days. Experiments were done at least thrice, with reproducible results.
Fluorescence microscopy
Calcofluor white (CFW, Fluorescent Brightener 28, Sigma) was used as a fluorochrome having excitation and emission wavelength of λ365 and λ435 respectively. Exponential cultures of yeast strains were fixed with 4% paraformaldehyde for 30 min at 30 °C with agitation (200 rpm). The cells were then washed twice with sterile phosphate-buffered saline (PBS), resuspended in PBS with 10 μg/ml CFW and incubated for 15 min at room temperature in the dark. Following staining cells were washed twice with PBS and resuspended in the same buffer, before observing with a fluorescence microscope using 100× objective lens. NIS-Elements AR 3.2 software was used for semi-quantitative analysis of fluorescence intensity. Individual cells from different frames for a sample were encircled and the mean fluorescence intensity (MFI) of each cell was obtained using analysis controls. In the case of pseudohyphae formation, individual cells were selected instead of whole filaments. Average fluorescence intensity for each sample was calculated from MFI of all the cells of that sample.
Zymolyase sensitivity assay
Zymolyase sensitivity of yeast strains was tested in pre-sterilized, polystyrene, flat-bottomed 96-well microtiter plates (Becton Dickinson), as described65, with some modifications. Yeast cells, equivalent to 0.5 OD600nm, were harvested from exponential culture, washed with PBS and resuspended in 200 μl zymolyase assay buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 4% PEG (8000) and 2 U (100 μg) of zymolyase 20 T (Seikagaku Corporation, Japan)65. The microtiter plate was incubated at 30 °C with shaking in microtiter plate reader (BioTek Microplate Reader; USA) and OD600nm was monitored for 90 min.
Hyphae formation assay
It was performed on YPD agar plate containing 10% fetal bovine serum (FBS) (Invitrogen). Five μl of normalised 1.0 OD600nm cells were spotted and incubated at 37 °C for 5 days before being photographed. To check invasive growth into agar, cells on the agar surface were washed away with water; the agar was then vertically sectioned and the agar slice was observed under a microscope with a 40× brightfield objective.
Biofilm formation and XTT reduction assay
Biofilms were formed in a 96-well microtiter plate as described previously66,67. For inoculum preparation, exponential YPD broth cultures were harvested, and cells were resuspended in RPMI-1640 medium at a density of 1 × 106 cells/ml. 100 μl of inoculums were dispensed into selected wells of 96-well microtiter plates and incubated at 37 °C for 2 days. After incubation, medium was gently aspirated from the wells and non-adherent cells were removed by washing thrice with sterile PBS. Residual PBS of wells was then removed by blotting with paper towels. Colorimetric XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium- 5-carboxanilide sodium salt] reduction assay was then performed for the quantification of biofilm formation as previously reported66,67. Briefly, 1 μM final concentration of menadione (Sigma; 10 mM prepared in acetone) was added to the filter sterilised (0.22 μm filter) stock solution of XTT tetrazolium salt (0.5 g/L) (Sigma). 100 μl of XTT-menadione solution was added into the prewashed preformed biofilms and to empty wells (for the background values of XTT reduction) of microtiter plates and incubated at 37 °C in the dark for 1 hr. Colorimetric change in the XTT reduction was measured in a microtiter plate reader at 492 nm.
Scanning electron microscopy (SEM)
For SEM, biofilms were formed on poly-L-lysine coated glass coverslips in 24-well cell culture plates (Nunc). 1 ml inoculums of 1 × 106 cells/ml were dispensed into selected wells of a microtiter plate and incubated at 37 °C for 2 days. After incubation, biofilms were processed and dried as described previously68, with some modifications. Briefly, preformed biofilms were washed 3-times with PBS and fixed subsequently for 20 min with formaldehyde (4% vol/vol) and glutaraldehyde (2% vol/vol). After fixation, biofilms were dehydrated through a series of ethanol solutions (30%, 50% and 70% for 10 min, 20 min and 30 min respectively at 4 °C and with 90% and 95% ethanol for 30 min at room temperature). Final dehydration was carried out by t-butyl alcohol for 30 min at room temperature and then dried in a desiccator. The samples were then coated with gold-palladium for 135 sec at 10–12 milliamps and observed with a scanning electron microscope (ZEISS EVO 40) in high-vacuum mode at 20 kV.
Amphotericin B susceptibility testing on preformed biofilms and during biofilm formation
For susceptibility assay on preformed biofilms, serially double-diluted AmB (0–16 μg/ml) in RPMI-1640 was dispensed (100 μl per well) into the wells of prewashed preformed biofilms and incubated at 37 °C for 2 days. For AmB susceptibility testing during biofilm formation, biofilms were formed in 96-well microtiter plates as described above with some modifications. For inoculums preparation, cells were resuspended in RPMI-1640 medium at a density of 2 × 106 cells/ml. Inoculums were dispensed (100 μl per well) in serially double-diluted concentrations of AmB (0–16 μg/ml), such that final cell density is 1 × 106 cells/ml for biofilm formation. At the end of incubation, AmB susceptibility of biofilm was measured by XTT reduction assay. AmB susceptibility experiments were performed on three different days in quadruplicates.
Additional Information
How to cite this article: Alfatah, M. et al. Critical role for CaFEN1 and CaFEN12 of Candida albicans in cell wall integrity and biofilm formation. Sci. Rep. 7, 40281; doi: 10.1038/srep40281 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Pfaller, M. & Diekema, D. Epidemiology of invasive candidiasis: a persistent public health problem. Clinical microbiology reviews 20, 133–163 (2007).
Sardi, J. C., Scorzoni, L., Bernardi, T., Fusco-Almeida, A. M. & Mendes Giannini, M. J. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 62, 10–24 (2013).
Andes, D. R. et al. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis 54, 1110–22 (2012).
Brown, G. D. et al. Hidden killers: human fungal infections. Sci Transl Med 4, 165rv13 (2012).
Denning, D. W. & Bromley, M. J. Infectious Disease. How to bolster the antifungal pipeline. Science 347, 1414–6 (2015).
Roemer, T. & Krysan, D. J. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb Perspect Med 4 (2014).
Anderson, T. M. et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol 10, 400–6 (2014).
Gray, K. C. et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci USA 109, 2234–9 (2012).
Delgado-Silva, Y. et al. Participation of Candida albicans Transcription Factor RLM1 in Cell Wall Biogenesis and Virulence. PloS One 9, e86270 (2014).
Chamilos, G. et al. Candida albicans Cas5, a regulator of cell wall integrity, is required for virulence in murine and toll mutant fly models. Journal of Infectious Diseases 200, 152–157 (2009).
Klis, F. M., Boorsma, A. & De Groot, P. W. Cell wall construction in Saccharomyces cerevisiae . Yeast 23, 185–202 (2006).
Lesage, G. & Bussey, H. Cell wall assembly in Saccharomyces cerevisiae . Microbiology and Molecular Biology Reviews 70, 317–343 (2006).
Cowen, L. E. & Steinbach, W. J. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryotic cell 7, 747–764 (2008).
Loeffler, J. & Stevens, D. A. Antifungal drug resistance. Clinical Infectious Diseases 36, S31–S41 (2003).
Smits, G. J., van den Ende, H. & Klis, F. M. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147, 781–794 (2001).
Reinoso-Martín, C., Schüller, C., Schuetzer-Muehlbauer, M. & Kuchler, K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryotic cell 2, 1200–1210 (2003).
Gerik, K. J., Bhimireddy, S. R., Ryerse, J. S., Specht, C. A. & Lodge, J. K. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans . Eukaryotic cell 7, 1685–1698 (2008).
Bermejo, C. et al. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Molecular biology of the cell 19, 1113–1124 (2008).
Levin, D. E. Cell wall integrity signaling in Saccharomyces cerevisiae . Microbiology and Molecular Biology Reviews 69, 262–291 (2005).
Roelants, F. M., Torrance, P. D., Bezman, N. & Thorner, J. Pkh1 and pkh2 differentially phosphorylate and activate ypk1 and ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Molecular biology of the cell 13, 3005–3028 (2002).
Dickson, R. C. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. Journal of lipid research 49, 909–921 (2008).
Sims, K. J., Spassieva, S. D., Voit, E. O. & Obeid, L. M. Yeast sphingolipid metabolism: clues and connections. Biochemistry and cell biology 82, 45–61 (2004).
Patton, J. L. & Lester, R. L. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. Journal of bacteriology 173, 3101–3108 (1991).
Ikonen, E. Cellular cholesterol trafficking and compartmentalization. Nature reviews molecular cell biology 9, 125–138 (2008).
Van Meer, G., Voelker, D. R. & Feigenson, G. W. Membrane lipids: where they are and how they behave. Nature reviews molecular cell biology 9, 112–124 (2008).
Simons, K. & Ikonen, E. Functional rafts in cell membranes. Nature 387, 569–72 (1997).
Lingwood, D. & Simons, K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010).
Ikonen, E. Roles of lipid rafts in membrane transport. Curr Opin Cell Biol 13, 470–7 (2001).
Mollinedo, F. Lipid raft involvement in yeast cell growth and death. Frontiers in oncology 2 (2012).
Simons, K. & Toomre, D. Lipid rafts and signal transduction. Nature reviews molecular cell biology 1, 31–39 (2000).
Brown, D. A. & London, E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol 14, 111–36 (1998).
Bagnat, M., Keranen, S., Shevchenko, A. & Simons, K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA 97, 3254–9 (2000).
Hanzal-Bayer, M. F. & Hancock, J. F. Lipid rafts and membrane traffic. FEBS letters 581, 2098–2104 (2007).
Simons, K. & Sampaio, J. L. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol 3, a004697 (2011).
Sharma, S. et al. Sphingolipid Biosynthetic Pathway Genes FEN1 and SUR4 Modulate Amphotericin B Resistance. Antimicrobial agents and chemotherapy 58, 2409–2414 (2014).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
Ram, A. F. & Klis, F. M. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nature protocols 1, 2253–2256 (2006).
Lussier, M. et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae . Genetics 147, 435–450 (1997).
Ragni, E. et al. The genetic interaction network of CCW12, a Saccharomyces cerevisiae gene required for cell wall integrity during budding and formation of mating projections. BMC genomics 12, 107 (2011).
Oh, C.-S., Toke, D. A., Mandala, S. & Martin, C. E. ELO2 and ELO3, Homologues of the Saccharomyces cerevisiae ELO1 Gene, Function in Fatty Acid Elongation and Are Required for Sphingolipid Formation. Journal of Biological Chemistry 272, 17376–17384 (1997).
Denic, V. & Weissman, J. S. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130, 663–677 (2007).
Ejsing, C. S. et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proceedings of the National Academy of Sciences 106, 2136–2141 (2009).
Popolo, L., Gilardelli, D., Bonfante, P. & Vai, M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae . Journal of bacteriology 179, 463–469 (1997).
Codlin, S., Haines, R. L., Jemima, J., Burden, E. & Mole, S. E. Btn1 affects cytokinesis and cell-wall deposition by independent mechanisms, one of which is linked to dysregulation of vacuole pH. Journal of cell science 121, 2860–2870 (2008).
Richard, M. et al. GPI7 affects cell-wall protein anchorage in Saccharomyces cerevisiae and Candida albicans . Microbiology 148, 2125–2133 (2002).
Revardel, E., Bonneau, M., Durrens, P. & Aigle, M. Characterization of a new gene family developing pleiotropic phenotypes upon mutation in Saccharomyces cerevisiae . Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 1263, 261–265 (1995).
Healey, K. R., Katiyar, S. K., Raj, S. & Edlind, T. D. CRS–MIS in Candida glabrata: sphingolipids modulate echinocandin–Fks interaction. Molecular microbiology 86, 303–313 (2012).
Healey, K. R., Challa, K. K., Edlind, T. D. & Katiyar, S. K. Sphingolipids mediate differential echinocandin susceptibility in Candida albicans and Aspergillus nidulans . Antimicrob Agents Chemother 59, 3377–84 (2015).
Sun, Y. et al. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Molecular and cellular biology 20, 4411–4419 (2000).
Ramanandraibe, E. et al. Implication of cell wall constituents in the sensitivity of Kluyveromyces lactis strains to amphotericin B. Research in microbiology 149, 109–118 (1998).
Seo, K., Akiyoshi, H. & Ohnishi, Y. Alteration of cell wall composition leads to amphotericin B resistance in Aspergillus flavus . Microbiology and immunology 43, 1017–1025 (1999).
Bahmed, K., Bonaly, R., Wathier, M., Pucci, B. & Coulon, J. Change of cell wall chitin content in amphotericin B resistant Kluyveromyces strains. FEMS microbiology letters 216, 99–103 (2002).
Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192, 775–818 (2012).
David, D., Sundarababu, S. & Gerst, J. E. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. The Journal of cell biology 143, 1167–1182 (1998).
Bari, V. K., Sharma, S., Alfatah, M., Mondal, A. K. & Ganesan, K. Plasma Membrane Proteolipid 3 Protein Modulates Amphotericin B Resistance through Sphingolipid Biosynthetic Pathway. Sci Rep 5, 9685 (2015).
Huang, Z. et al. A functional variomics tool for discovering drug-resistance genes and drug targets. Cell Rep 3, 577–85 (2013).
Finkel, J. S. & Mitchell, A. P. Genetic control of Candida albicans biofilm development. Nature Reviews Microbiology 9, 109–118 (2010).
Sudbery, P. E. Growth of Candida albicans hyphae. Nature Reviews Microbiology 9, 737–748 (2011).
Brand, A. Hyphal growth in human fungal pathogens and its role in virulence. International journal of microbiology 2012 (2011).
Mayer, F. L., Wilson, D. & Hube, B. Candida albicans pathogenicity mechanisms. Virulence 4, 119 (2013).
Mathe, L. & Van Dijck, P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet 59, 251–64 (2013).
Nett, J. E. Future directions for anti-biofilm therapeutics targeting Candida . Expert review of anti-infective therapy 12, 375–382 (2014).
Walraven, C. J. & Lee, S. A. Antifungal lock therapy. Antimicrob Agents Chemother 57, 1–8 (2013).
Lattif, A. A. et al. Lipidomics of Candida albicans biofilms reveals phase-dependent production of phospholipid molecular classes and role for lipid rafts in biofilm formation. Microbiology 157, 3232–3242 (2011).
Puria, R., Mannan, M., Chopra‐Dewasthaly, R. & Ganesan, K. Critical role of RPI1 in the stress tolerance of yeast during ethanolic fermentation. FEMS yeast research 9, 1161–1171 (2009).
Nett, J. E., Cain, M. T., Crawford, K. & Andes, D. R. Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. Journal of Clinical Microbiology 49, 1426–1433 (2011).
Ramage, G., Walle, K. V., Wickes, B. L. & López-Ribot, J. L. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrobial agents and chemotherapy 45, 2475–2479 (2001).
Ramage, G., Saville, S. P., Wickes, B. L. & López-Ribot, J. L. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Applied and Environmental Microbiology 68, 5459–5463 (2002).
Winston, F., Dollard, C. & Ricupero‐Hovasse, S. L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55 (1995).
Baker Brachmann, C. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998).
Fonzi, W. A. & Irwin, M. Y. Isogenic strain construction and gene mapping in Candida albicans . Genetics 134, 717–728 (1993).
Noble, S. M. & Johnson, A. D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans . Eukaryotic cell 4, 298–309 (2005).
Acknowledgements
Md. Alfatah, Vinay K. Bari, Anubhav S. Nahar and Swati Bijlani acknowledge the Council of Scientific and Industrial Research, New Delhi and University Grants Commission, New Delhi, for fellowships. We also acknowledge the help of Anil Theophilus for Scanning electron microscopy and Deepak Bhatt for confocal microscopy and image analysis. This work was supported by CSIR projects on “Infectious Diseases” (BSC0210) and “Bugs to Drugs” (BSC0211).
Author information
Authors and Affiliations
Contributions
K.G. designed the project and provided overall guidance. M.A., V.K.B., A.S.N. and S.B. carried out the experiments and collected data. M.A. and K.G. drafted and finalised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Alfatah, M., Bari, V., Nahar, A. et al. Critical role for CaFEN1 and CaFEN12 of Candida albicans in cell wall integrity and biofilm formation. Sci Rep 7, 40281 (2017). https://doi.org/10.1038/srep40281
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40281
This article is cited by
-
Candida albicans gains azole resistance by altering sphingolipid composition
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.