Abstract
The CRISPR/Cas system is an efficient genome-editing tool to modify genes in mouse zygotes. However, only the Streptococcus pyogenes Cas9 (SpCas9) has been systematically tested for generating gene-modified mice. The protospacer adjacent motif (PAM, 5′-NGG-3′) recognized by SpCas9 limits the number of potential target sites for this system. Staphylococcus aureus Cas9 (SaCas9), with its smaller size and unique PAM (5′-NNGRRT-3′) preferences, presents an alternative for genome editing in zygotes. Here, we showed that SaCas9 could efficiently and specifically edit the X-linked gene Slx2 and the autosomal gene Zp1 in mouse zygotes. SaCas9-mediated disruption of the tyrosinase (Tyr) gene led to C57BL/6J mice with mosaic coat color. Furthermore, multiplex targeting proved efficient multiple genes disruption when we co-injected gRNAs targeting Slx2, Zp1, and Tyr together with SaCas9 mRNA. We were also able to insert a Flag tag at the C-terminus of histone H1c, when a Flag-encoding single-stranded DNA oligo was co-introduced into mouse zygotes with SaCas9 mRNA and the gRNA. These results indicate that SaCas9 can specifically cleave the target gene locus, leading to successful gene knock-out and precise knock-in in mouse zygotes, and highlight the potential of using SaCas9 for genome editing in preimplantation embryos and producing gene-modified animal models.
Similar content being viewed by others
Introduction
Gene-modified mouse models are important tools in understanding gene function in vivo and developing new therapeutics for diseases. Before the advent of nuclease-based genome-editing tools, gene-modified mice were generated mainly by gene targeting in embryonic stem cells (ESCs), a time-consuming and labour-intensive process1. Several classes of artificial nucleases have been developed, including zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and the CRISPR/Cas system2,3. These nucleases have been shown to work efficiently in zygotes from different species, and not restricted to ESCs, thus enabling one-step generation of gene-modified animals with drastically improved efficiency and reduced cost and time commitment4,5,6,7,8,9,10,11,12. In zygotes, the nucleases induce double-strand breaks (DSBs) at target sites, which are subsequently repaired through either non-homologous end joining (NHEJ) or homology-directed repair (HDR), resulting in possible frame-shift mutations and precise gene modifications, respectively2,13. ZFN and TALEN are bipartite fusion proteins consisting of nuclease domains as well as DNA-binding domains that recognize the target site by protein-DNA interaction2. Complex protein engineering steps are required to generate site-specific ZFN and TALEN, which has greatly hampered the application and expansion of these platforms3.
The CRISPR/Cas system, which consists of the Cas nuclease and guide RNA (gRNA), is adapted from the acquired immune systems of bacteria and archaea14,15,16. The 20-nucleotide sequence at the 5′ or 3′ end of the gRNA specifies the target site through RNA-DNA hybridization17,18,19. As a result, targeting different genes using CRISPR/Cas can be more easily achieved through the manipulation of gRNA sequences. Its simplicity, flexibility, and high efficiency has enabled the application of CRISPR/Cas systems in generating a variety of gene-modified species within a short amount of time4,6,10,11,17,18,19,20,21,22,23,24. Of the different CRISPR/Cas systems, such as Streptococcus pyogenes Cas9 (SpCas9), Staphylococcus aureus Cas9 (SaCas9), Lachnospiraceae Cpf1 (LbCpf1), Acidaminococcus Cpf1 (AsCpf1), Streptococcus thermophilus Cas9 (StCas9), and Neisseria meningitidis Cas9 (NmCas9), only SpCas9 has been extensively characterized and widely used in animal zygotes genome editing6,7,20,21,22,24,25,26,27. However, its specific protospacer adjacent motif (PAM, 5′-NGG-3′) preference limits the number of potential genome recognition sites for SpCas9, which calls for alternative CRISPR/Cas systems for genome editing in zygotes.
SaCas9 is derived from Staphylococcus aureus, pathogenic bacteria that can cause human skin and soft tissue infections28,29,30. SaCas9 is the smallest among all the known Cas nucleases, largely due to its small recognition (REC) lobe28. This small size makes it particular attractive for gene therapy, which often involves in vivo delivery using viral vectors. In addition, SaCas9 recognizes a unique 5′-NNGRRT-3′ PAM sequence through its PAM-interacting (PI) domain29. Moreover, the PAM sequence of SaCas9 is predicted to occur once every 32 bps, compared to once every 8 bps for SpCas9. These unique properties suggest specific recognition and cleavage by SaCas9, which is supported by the genome-wide sequencing data reported recently31. Despite its promise in genome editing, no reports to date have examined the ability of SaCas9 to modify genomes in zygotes.
In this report, we compared the genome editing efficiency and specificity of SaCas9 vs. SpCas9 in mouse zygotes, and examined the resultant mosaicism. In addition, we also provided evidence that demonstrated the feasibility of using SaCas9 for multiplex gene disruption and homology-directed repair in mouse embryos. Our data reveal that SaCas9 is comparable to SpCas9 in genome editing in animal zygotes, and highlight the potential of SaCas9 as an alternative to SpCas9 for modifying genomes.
Results
SaCas9 cleaves target sites efficiently in mouse embryos
To test whether SaCas9 could edit genes in mouse zygotes, we designed two Staphylococcus aureus gRNAs (Sa-gRNAs) to respectively target the X-linked gene Slx2 and autosomal gene Zp1 (Fig. 1A). For comparison, Streptococcus pyogenes gRNAs (Sp-gRNAs) which could also target Slx2 and Zp1 were designed as well (Fig. 1A). Next, we injected the gRNAs with the mRNA of their respective Cas9 nucleases into 0.5-day mouse zygotes. The injected embryos were either transferred into 0.5-day pseudopregnant mice 2 hours after microinjection or cultured in vitro and genotyped. The cultured embryos were harvested 48 hours later to PCR amplify the target regions for the T7 endonuclease I (T7EI) assay and Sanger sequencing (Fig. 1B). As shown in Fig. 1C, the T7EI assay indicated that SaCas9 was able to cleave both the Slx2 and Zp1 loci efficiently, at levels comparable to SpCas9. Sanger sequencing of the PCR amplicons further revealed that 24/27 (88.8%) of the embryos had been cleaved by Slx2-SaCas9, compared to 14/26 (53.8%) for Slx2-SpCas9, and 23/25 (92%) of the embryos cleaved by Zp1-SaCas9 vs. 26/27 (96.3%) by Zp1-SpCas9 (Fig. 1D and Table 1). These results demonstrate the feasibility of using SaCas9 to edit target genes in mouse zygotes.
(A) Schematic representations of the Slx2 and Zp1 target sites. Here, SaCas9 and SpCas9 target the same regions of Slx2 and Zp1, their target sequences are underlined respectively in red and blue. PAM sequences are underlined in green. (B) A flow chart indicating the strategy to determine the cleavage efficiencies of SaCas9. SpCas9 was used for comparsion. Cas9 mRNAs and their respective gRNAs were injected into 0.5-day old zygotes. After 48 hours, a portion of the injected embryos were in vitro cultured for the T7 endonuclease I (T7EI) assay and Sanger sequencing. (C) The regions spanning the target sites of the Slx2 and Zp1 loci were PCR amplified. The PCR products were then used for the T7EI assay. Black arrowheads indicate PCR fragments that contain no mismatch. WT, wild-type. (D) The PCR products from (C) were also examined by Sanger sequencing. Representative sequencing results of SaCas9 or SpCas9-cleaved embryos are presented here. The presences of multiple peaks near the PAM sequences indicate sites of cleavage and NHEJ-mediated repair.
Efficient generation of mutant mice by SaCas9
Following our in vitro analysis of SaCas9-mediated genome editing in embryos, we went on to evaluate the efficiency and specificity of SaCas9 in mutant mice. The birth rates of mice 20 days after the transplantation of SaCas9 and SpCas9 modified embryos were calculated and shown in Table 1, where comparable birth rates of SaCas9 vs. SpCas9 were found (17.9% and 8.3% compared to 9.5% and 16.5%). This result indicates low embryo toxicity for SaCas9. Preliminary T7EI analysis of the pups indicated the presence of mutant mice as a result of cleavage by SaCas9 or SpCas9 (Fig. 2A). Further verification by Sanger sequencing showed that some of pups that had been deemed wild-type by the T7EI assay were actually homozygous mutant mice (Fig. 2B). As a result, all pups were verified by Sanger sequencing to more accurately estimate Cas9 efficiency. Again, SaCas9 appeared comparable to SpCas9, 16/17 (94.1%) pups were edited by SaCas9 as opposed to 8/17 (47.1%) by SpCas9 for the Slx2 locus, and 14/18 (77.7%) vs. 10/12 (83.3%) for the Zp1 locus (Table 1). These results suggest that as has been found for SpCas9, the efficiency of SaCas9 is site dependent and that the overall efficiency of SaCas9 is similar to that of SpCas96.
The injected embryos from Fig. 1 were transplanted into pseudopregnant mice to generate mutant mice. (A) PCR amplification and T7EI assays were carried out as described above to identify mutant founder mice. Arrowheads denote uncut PCR products with no mismatch. *Indicates pups that appeared to be wild-type in the T7EI assay, but turned out to be homozygous mutants when analysed by Sanger sequencing. (B) Representative sequencing chromatographs are presented here, with the target sequences (PAM in green) shown above. Most mutant mice resemble pup #5 from the Zp1-SaCas9 group, where multiple peaks are present near the PAM sequence. Others resemble pup #2 from the Slx2-SaCas9 group, #4 from Slx2-SpCas9, and #6 from Zp1-SpCas9, in which deletions of various sizes (1–10 bps) were apparent. These are the same samples that were deemed uncut (wild-type) in the T7EI assay in (A). Red arrowheads indicate sites with base pair deletion, with the deleted residues underlined in red. (C) Ten pups were randomly selected from each group. Regions encompassing the top ten predicted off-target sites (numbered on the x-axis) were PCR amplified using the genomic DNA from these pups and analysed by deep sequencing. The percentages of modifications at each site, including the intended target, are plotted on the y-axis. Only 9 samples from Slx2-SaCas9 were plotted here because of insufficient sequencing depth in one sample. Of the Slx2-SpCas9 group, one supposedly “wild-type” pup was actually cleaved by SpCas9 at a very low frequency, while the other one was not cleaved at all. For both Slx2-SaCas9 and Slx2-SpCas9, the cleavage efficiency appeared low in several samples.
The unique 5′-NNGRRT-3′ PAM preference of SaCas9 should in theory lead to fewer off-target sites than SpCas9. To further compare the specificity of SaCas9 and SpCas9 in founder mice, we selected 10 mutant pups from each group, except that eight mutant and two wild-type (WT) pups (verified by Sanger sequencing) (Table 1) were chosen for the Slx2-SpCas9 group. On-target sites of Slx2 and Zp1 as well as the top ten potential off-target sites were PCR amplified and deep sequenced (Fig. 2C). None of the potential off-target sites examined appeared to be cleaved by either SaCas9 or SpCas9, suggesting that both Cas9 nucleases are very specific. It is interesting to note that deep sequencing revealed very low gene modification efficiency (1.71%) in one pup, a “wild-type” mouse in the Slx2-SpCas9 group as verified by Sanger sequencing, likely because deep sequencing is much more sensitive than Sanger sequencing at detecting indels.
Mutant founder mice generated by SaCas9 are mosaic
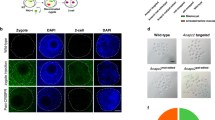
One limitation of using SpCas9 in generating gene-modified mice is that most of the founder mice are mosaic, which complicates phenotypic analysis of founder mice. To determine whether SaCas9 had similar issues, we designed 2 Sa-gRNAs (SaCas9-G1 and SaCas9-G2) and 2 Sp-gRNAs (SpCas9-G3 and SpCas9-G4) to target exon 4 of the tyrosinase gene (Tyr) (Fig. 3A). Tyrosinase is a key enzyme for melanin production whose dysfunction leads to albinism in C57BL/6J mice. Albinism in C57BL/6J mice makes it easy to visualize mosaicism in the offspring32. Most of the injected zygotes were transferred into pseudopregnant mothers. We genotyped 8 embryos using the T7EI assay after 48 hours of in vitro culture and found that both SaCas9 and SpCas9 could edit the Tyr gene (Fig. 3B). About 9 days after birth, both mosaic coloured and totally albino pups could be found in SaCas9 and SpCas9 groups (Fig. 3C). These albino and mosaic pups were confirmed to be C57 by microsatellite analysis (Supplementary information, Fig. S1). For SpCas9, the percentages of albino pups were 66.7% and 68.8% (Table 2). For SaCas9, the albino rate was 72.7% for SaCas9-G1, but 38.1% for SaCas9-G2. These data indicate that SaCas9-generated founder mice are mosaic, in agreement with the sequencing results of Tyr loci in the pups (Supplementary information, Fig. S2).
(A) A schematic representation of the target sites in the tyrosinase (Tyr) gene. The SaCas9 and SpCas9 target sequences are respectively underlined in red and blue, with PAM sequences underlined in green. (B) T7EI analysis of the injected embryos was performed to determine the cleavage efficiency of SaCas9 and SpCas9 at the Tyr locus. Arrowheads indicate uncut (or no mismatch) PCR products. WT, wild-type. (C) Coat colour was assessed in founder mice nine days after birth. Albino and mosaic (yellow*) founders were apparent in mice obtained from embyros modified by SaCas9 or SpCas9. (D) Genomic DNA was extracted from newborn mice and used in T7EI assays to assess Cas9 cleavage at the Tyr locus. Arrowheads indicate uncut (or no mismatch) PCR products. *Indicates pups that appeared to be wild-type in the T7EI assay, but turned out to be homozygous mutants when analysed by Sanger sequencing.
To further compare the efficiency of SaCas9 and SpCas9-mediated mutant mice generation, the genotypes of these pups were examined by both T7EI assay and Sanger sequencing (Fig. 3D, and Supplementary information, Fig. S3), which found that 3 black pups had actually been cleaved by SaCas9 at the Tyr locus (Table 2). The efficiency of SaCas9-G1 and SaCas9-G2 were 81.8% and 61.9%, respectively, comparable to that of SpCas9-G3 (66.7%) and SpCas9-G4 (81.3%) (Table 2). Again, these data underline the site dependence of the genome editing efficiency of SaCas9.
Multiplex gene disruption in mouse embryos by SaCas9
Our data thus far indicated that SaCas9 can cleave efficiently in a single gene locus in mouse embryos. We next addressed whether SaCas9 could disrupt multiple genes simultaneously. SaCas9 mRNA and multiple gRNAs that target different loci (Slx2-SaCas9, Zp1-SaCas9, and Tyr-SaCas9-G1) were co-injected into the cytoplasm of mouse zygotes. SpCas9 mRNA and the corresponding SpCas9 gRNAs (Slx2-SpCas9, Zp1-SpCas9 and Tyr-SpCas9-G3) were also injected into zygotes as a control. The injected embryos were harvested 48 hours later for whole-genome amplification by multiplex displacement amplification (MDA) and the target regions were PCR amplified and genotyped (Fig. 4A and Supplementary information, Fig. S4). Five embryos from the SaCas9 group (5/9, 55.6%) had been targeted at all three loci. In contrast, only 2/9 (22.2%) embryos from the SpCas9 group were triple targeted (Fig. 4A). The genotypes, including single- and double-targeted embryos, are summarized in Table 3. Our data revealed that 46.7% (14/30) of SaCas9-injected embryos were triple targeted, while only 8.1% (3/37) of SpCas9-injected embryos were triple-targeted (Table 3 and Supplementary information, Table S1). These results demonstrate the feasibility of multiplex gene disruption using SaCas9.
(A) gRNAs targeting Tyr, Zp1, and Slx2 were simultaneously injected into mouse zygotes with their respective Cas9 mRNAs. Regions spanning the target sites were PCR amplified for T7EI analysis. Arrowheads indicate uncut (or no mismatch) PCR products. All embryos were further genotyped by Sanger sequencing. *Indicates triple targeted embryos. Sanger sequencing revealed that #2, #3, #6 and #8 embryos from Zp1 and #8 embryo from Slx2 (both in the SaCas9 group) had homozygous deletions. And Tyr and Zp1 from #8 embryos of SpCas9 group also had homozygous deletions. (B) The design of the gRNA and single-stranded DNA oligo for the H1c locus. The target site is underlined in red, and the PAM sequence in green. The 2 × Flag tag coding sequence is in green and StuI site in red. (C) The region encompassing the target site of the H1c locus was PCR amplified and then digested with StuI for RFLP (Restriction fragment length polymorphism) analysis. *Indicates embryos repaired by HDR. (D) PCR products from (C) were TA cloned for sequence verification of successful knock-in. A Representative sequencing chromatograph is shown. The 2 × Flag tag sequence and StuI site are underlined respectively in green and red.
Precise gene knock-in modification in mouse embryos mediated by SaCas9
Our results clearly show that SaCas9 can generate indels via the NHEJ pathway, which leads to gene knockout (KO). Next, we explored the possibility of using SaCas9 to generate knock-in (KI) mice, which requires a donor template and utilizes the HDR mechanism. We designed a Sa-gRNA to target the 3′ end of the histone H1c coding sequence, and a single-stranded DNA oligo encoding a 2 × Flag tag and a StuI restriction site flanked by homology arms (Fig. 4B). We co-injected the DNA oligo and gRNA together with SaCas9 mRNA into zygotes and collected the embryos after 48 hours to PCR amplify the H1c target region. If a PCR product could be digested with StuI, it would indicate successful insertion of the oligo sequence. Indeed, 2 out of 12 embryos appeared to have incorporated the StuI site (Fig. 4C). The PCR products were subsequently TA cloned for Sanger sequencing (Fig. 4D). We found that sequences for the 2 × Flag tag as well as the StuI site were both integrated into the H1c gene locus, supporting the notion that SaCas9 can efficiently mediate gene knock-in through the HDR mechanism.
Discussion
The rapid development and application of CRISPR/Cas9 technology has led to numerous studies on its efficacy and specificity, although the majority of studies have focused on SpCas9. Here, we describe the first attempt to systematically determine the genome editing efficiency and specificity of SaCas9 in mouse embryos. Using multiple assays and platforms, we showed that the efficiency of SaCas9 was comparable to that of SpCas9. This finding is in agreement with results reported for human cell lines33. Three independent studies have found SaCas9 to be specific at different target sites in human cells29,31,33. Our deep sequencing results also support high specificity for SaCas9 in mouse embryos. Multiplex gene disruption and one-step knock-in are some of the key advantages of SpCas9. By simultaneously targeting three genes, we demonstrated that SaCas9 was also capable of multiplex gene disruption. And we successfully tagged endogenous histone H1c on the c-terminus with a 2 × Flag tag. It has been well documented, although not fully understood, that Cas9/gRNA specificity often depends on the particular sequences and/or structures of the regions being targeted. Therefore, extensive high-throughput sequencing investigations (e.g., using GUIDE-seq or BLESS) are sorely needed to better elucidate the exact factors that determine the specificity of SaCas9 and SpCas9 at different genomic sites. And the efficiency of SaCas9-mediated multiplex gene disruption and gene knock-in in comparison with SpCas9 warrants further investigation. Mosaicism has been a common phenomenon in SpCas9-generated founder mice, where an animal is a chimera of both wild-type and mutant cells3,34. It is mainly caused by inefficient cleavage of the target site in one-cell stage embryos and prolonged nuclease activities following the first round of cell division34. Whether SaCas9 could also lead to mosaicism had been largely unknown. By targeting the coat-colour gene tyrosinase (Tyr), we found that SaCas9-generated founder mice also exhibited mosaicism, similar to those generated by SpCas932.
Our work on SaCas9 in mouse models indicates that SaCas9, similar to SpCas9, can be harnessed to edit genomes of other animals such as rats, pigs, and monkeys4,10,20,22,23,24,35. Dr. Yong Fan’s group and ours have shown that SpCas9 could edit CCR5 andβ-globin genes in human tripronuclear zygotes36,37. The UK Human Fertilisation and Embryology Authority (HFEA) recently licensed Dr. Kathy Niakan to edit human embryo genomes for the mechanistic study of human preimplantation embryos38. SaCas9 may be an attractive alternative to SpCas9 in similar studies. Indeed, experiments have been carried out to optimize SaCas9 and expand its applications. The structure and catalytic residues of SaCas9 have been reported, and nuclease-dead SaCas9 has been created28. When fused with the KRAB (Krüppel-associated box) domain of KOX1, VP16, the histone acetyltransferase (HAT) core domain of P300, or the DNA methylation domain of DNMT3A, nuclease-dead SaCas9 may enable precise transcriptional and epigenetic regulation28,39,40,41,42. Dr. J. Keith Joung and colleagues reported an engineered SaCas9 protein that recognized the 5′-NNNRRT-3′ PAM sequence, potentially broadening the targetable sites of SaCas931. Dr. Feng Zhang’s group developed a SaCas9 variant with higher specificity by replacing several positively charged residues with uncharged amino acids43. Continued improvements should greatly enhance our ability to use SaCas9 in research, broaden its applications in other fields, and facilitate potential clinical implementations of CRISPR/Cas9.
Materials and Methods
Ethics Statement
All experiments were conducted with protocols approved by The Institutional Animal Care and Use Committee at Sun Yat-sen University.
Animal Experiments
Mice were purchased from Guangdong Medical Laboratory Animal Center. All mice were housed under climate-controlled conditions (22 ± 1 °C) in a specific pathogen-free animal facility with 14-hour light/10-hour dark cycles in Sun Yat-sen University. C57BL/6J mice (6–8 weeks old females) were superovulated and mated with C57BL/6J males. Plugged females were sacrificed by cervical dislocation. 0.5-day zygotes were collected using potassium simplex optimized medium (KSOM) containing N-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid and sodium bicarbonate (HKSOM), and cultured in KSOM until genotyping or transplantation. The embryos were in vitro cultured for 48 hours before genotyping or whole genome amplification. CD1 female mice (6–8 weeks old) that were mated with sterilized CD1 male mice were used as foster mothers.
In vitro transcription of Cas9 mRNA and gRNA
The pT7-3 × Flag-hCas9 plasmid was constructed as reported previously37. The transcription template of SaCas9 mRNA was PCR amplified from pX601 (Addgene) using the primer listed in Supplementary information, Table S2. We added the T7 promoter sequence to the 5′ end of the SaCas9 forward primer. The transcription templates were then purified using the PCR Clean Up Kit (Qiagen). SpCas9 and SaCas9 were transcribed using the mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies) following the manufacturer’s instruction. Sequences for SpCas9 gRNAs were cloned into the pDR274 vector (Addgene), using primers listed in Supplementary information Table S2. These recombinant vectors were then linearized with Dra I (NEB) and transcribed using the MEGAshortscript T7 kit (Life Technologies) following the manufacturer’s instruction. The transcription templates of SaCas9 gRNAs were PCR amplified from pX601 (Addgene) using primers listed in Supplementary information, Table S2. We also added the T7 promoter sequence to the 5′ end of the gRNA forward primer. The gRNAs were then transcribed using the MEGAshortscript T7 kit (Life Technologies). Cas9 mRNAs and the gRNAs were subsequently purified using the MEGAclear kit (Life Technologies) and resuspended in RNase-free water.
Intracytoplasmic injection of Cas9 mRNA and gRNA
The mixture of Cas9 mRNA (200 ng/μl) and gRNA (100 ng/μl) was injected into 0.5-day zygotes of C57BL/6J mice. About 2 hours after injection, the injected zygotes were transplanted into the oviduct of 0.5-day pseudopregnant mothers. Primers used for in vitro transcription of Cas9 mRNA and gRNA are listed in Supplementary information, Table S2.
Single embryo PCR amplification and mouse genotyping
Single embryo PCR amplification was performed as described by Ran et al.44. Briefly, each embryo was transferred into a PCR tube containing 1 μl lysis buffer, and then incubated at 65 °C for 3 hours followed by 95 °C for 10 min. The lysis product was then amplified using primers listed in Supplementary information, Table S3. Mouse genotyping was done by PCR and sequencing of tail-snips using the Mouse Genotyping Kit (KAPA Biosystems) following the manufacturer’s instruction. Primers used for genotyping are listed in Supplementary information, Table S3.
Genomic DNA analysis
Target sites were PCR amplified using primers listed in Supplementary information, Table S3. The PCR products were then used in the T7 endonuclease I (T7EI) cleavage assay as described by Shen et al.45. Primers for direct sequencing of the PCR products, which reveals the presence of double peaks or indels, are listed in Supplementary information, Table S3. PCR products with double peaks were then TA cloned into the pGEM-T vector (Promega) for plasmid DNA extraction and Sanger sequencing.
Microsatellite analysis
The D7Mit145 site and D12Mit136 site were amplified from albino and mosaic founders. The PCR products were then resolved by electrophoresis on a 10% PAGE gel. Information of primers were in Supplementary information, Table S4.
Whole genome amplification of embryonic genome DNA
Whole genome amplification of the embryos was performed using the PEPLI-g Midi Kit (Qiagen). Briefly, embryos were transferred into PCR tubes containing reconstituted buffer D2 (7 μl), and then incubated at 65 °C for 10 min. Then STOP solution (3.5 μl) was added to the reaction mixture, followed by addition of the MDA master mix (40 μl) and incubation at 30 °C for 8 hours. The reaction product was diluted with ddH2O (3:100), and 1 μl of the diluted DNA was used for PCR amplification. PCR products were then analyzed by the T7EI assay and Sanger sequencing.
Analysis of off-target sites by deep sequencing
We used the online tool Cas-OFFinder (http://www.rgenome.net/cas-offinder/) to identify potential off-target sites. The mismatch number and bulge size values are: Slx2-SpCas9, 4 and 0 respectively; Slx2-SaCas9, 5 and 0 respectively; Zp1-SpCas9, 2 and 0 respectively; and Zp1-SaCas9, 6 and 0 respectively. Sequences surrounding these sites were PCR amplified and deep sequenced using Illumina Hiseq 2500 PE250 as paired-end 250 reads. The primers for off-target effect analysis are listed in Supplementary information, Table S5. High-throughput sequencing data were analyzed as previously reported46. Briefly, sequencing reads were mapped to the mouse reference genome (UCSC, mm10) by means of BWA with default parameters (v0.7.8). Samtools and Picard tools were used to build index and sort mapped reads by GATK (Genome Analysis ToolKit, version 3.1–1). HaplotypeCaller was used to call variants and each then was divided into indels and SNVs in virtue of SelectVariants. We then excluded indels in the flanking region (±20bp) that did not overlap with the predicted on- and off-target sites by Bedtools. Indel site and indel frequency were calculated. We included 1-bp indel mutations only if they occurred directly adjacent to the cleavage site as reported46.
Additional Information
How to cite this article: Zhang, X. et al. Efficient Production of Gene-Modified Mice using Staphylococcus aureus Cas9. Sci. Rep. 6, 32565; doi: 10.1038/srep32565 (2016).
References
Capecchi, M. R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet 6, 507–512 (2005).
Gaj, T., Gersbach, C. A. & Barbas, C. F. 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31, 397–405 (2013).
Kim, H. & Kim, J. S. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15, 321–334 (2014).
Hai, T., Teng, F., Guo, R., Li, W. & Zhou, Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24, 372–375 (2014).
Carlson, D. F. et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA 109, 17382–17387 (2012).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Meyer, M., de Angelis, M. H., Wurst, W. & Kuhn, R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA 107, 15022–15026 (2010).
Qiu, Z. et al. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res 41, e120 (2013).
Niu, Y. et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843 (2014).
Ma, Y. et al. Generating rats with conditional alleles using CRISPR/Cas9. Cell Res 24, 122–125 (2014).
Long, C. et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science (2014).
Sander, J. D. & Joung, J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32, 347–355 (2014).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11, 181–190 (2010).
Horvath, P. & Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 (2010).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Li, D. et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31, 681–683 (2013).
Hwang, W. Y. et al. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 8, e68708 (2013).
Li, W., Teng, F., Li, T. & Zhou, Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 31, 684–686 (2013).
Chen, Y. et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet 24, 3764–3774 (2015).
Kang, Y. et al. CRISPR/Cas9-mediated Dax1 knockout in the monkey recapitulates human AHC-HH. Human molecular genetics 24, 7255–7264 (2015).
Zetsche, B. et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759–771 (2015).
Hou, Z. et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA 110, 15644–15649 (2013).
Chen, Y. et al. Functional disruption of the dystrophin gene in Rhesus Monkey Using CRISPR/Cas9. Hum Mol Genet (2015).
Nishimasu, H. et al. Crystal Structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Miller, L. G. et al. Staphylococcus aureus skin infection recurrences among household members: an examination of host, behavioral, and pathogen-level predictors. Clin Infect Dis 60, 753–763 (2015).
Kleinstiver, B. P. et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol (2015).
Yen, S. T. et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol 393, 3–9 (2014).
Friedland, A. E. et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol 16, 257 (2015).
Singh, P., Schimenti, J. C. & Bolcun-Filas, E. A mouse geneticist’s practical guide to CRISPR applications. Genetics 199, 1–15 (2015).
Wan, H. et al. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res 25, 258–261 (2015).
Kang, X. et al. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J Assist Reprod Genet (2016).
Liang, P. et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein & cell 6, 363–372 (2015).
Callaway, E. UK scientists gain licence to edit genes in human embryos. Nature 530, 18 (2016).
Hilton, I. B. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33, 510–517 (2015).
Cheng, A. W. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23, 1163–1171 (2013).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Vojta, A. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res (2016).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science (2015).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Shen, B. et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23, 720–723 (2013).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature (2016).
Acknowledgements
We thank Dr. Dan Liu for critical reading of the manuscript. This study was supported by the Science and Technology Planning Project of Guangdong Province (2015B020228002 and 2015A020212005), the Natural Science Foundation of Guangdong Province (2014A030312011 and 2016A030310206), the National Natural Science Foundation (31371508 and 81330055), the National Basic Research Program of China (973 Program, 2012CB947604), the Program of International S&T Cooperation (2014DFB30020), and the Fundamental Research Funds for the Central Universities (161gzd13 and 161gpy32).
Author information
Authors and Affiliations
Contributions
J.H. and Z.S. conceived the project and were involved in experimental design. X.Z., P.L., C.D., Z.Z., J.Z., X.X., R.H., Y.S., H.S., J.Z. and Y.X. performed experiments. X.Z., P.L. and C.D. analyzed the data. X.Z., P.L. and J.H. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, X., Liang, P., Ding, C. et al. Efficient Production of Gene-Modified Mice using Staphylococcus aureus Cas9. Sci Rep 6, 32565 (2016). https://doi.org/10.1038/srep32565
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32565
This article is cited by
-
Targeted disruption of tyrosinase causes melanin reduction in Carassius auratus cuvieri and its hybrid progeny
Science China Life Sciences (2019)
-
Effective gene editing by high-fidelity base editor 2 in mouse zygotes
Protein & Cell (2017)
-
Correction of β-thalassemia mutant by base editor in human embryos
Protein & Cell (2017)
-
CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools
Mammalian Genome (2017)
-
Efficient genome editing of differentiated renal epithelial cells
Pflügers Archiv - European Journal of Physiology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.