Abstract
The field of microbiology was established in the 17th century upon the discovery of microorganisms by Antonie van Leeuwenhoek using a single-lens microscope. Now, the detailed ultrastructures of microorganisms can be elucidated in situ using three-dimensional electron microscopy. Since the availability of electron microscopy, the taxonomy of microscopic organisms has entered a new era. Here, we established a new taxonomic system of the primitive algal genus Glaucocystis (Glaucophyta) using a new-generation electron microscopic methodology: ultra-high-voltage electron microscopy (UHVEM) and field-emission scanning electron microscopy (FE-SEM). Various globally distributed Glaucocystis strains were delineated into six species, based on differences in in situ ultrastructural features of the protoplast periphery under UHVEM tomography and in the mother cell wall by FE-SEM, as well as differences in the light microscopic characteristics and molecular phylogenetic results. The present work on Glaucocystis provides a model case of new-generation taxonomy.
Similar content being viewed by others
Introduction
Scientists were unaware of the existence of microorganisms until their discovery in 1674 by Antonie van Leeuwenhoek using a single-lens microscope, thereby establishing the field of microbiology1,2. Electron microscopy (EM), developed in the 20th century, has also contributed much to our understanding of microorganism ultrastructural characteristics3,4,5. However, morphological delineation of species in unicellular or colonial organisms has been limited compared with that in macroorganisms, especially in terms of their three-dimensional (3D) characteristics6,7.
Glaucophytes constitute one major lineage of such microorganisms. They are rare freshwater algae retaining the most ancestral features of primary photosynthetic eukaryotes or Archaeplastida7, which also include red algae and Chloroplastida (green algae and land plants7). Thus, glaucophyte algae represent an evolutionarily important group within Archaeplastida. However, species concepts of glaucophytes have fallen behind those in other archaeplastidal lineages, because the ability to determine morphological differences by light microscopy (LM) and conventional EM is limited8,9,10 (see Supplementary Note). Schnepf et al.8 observed three strains of the glaucophyte genus Glaucocystis (SAG 229-1, SAG 229-2 and SAG 229-3) by ultrathin-section transmission electron microscopy (TEM) and reported no ultrastructural differences among them. However, since their observations, no comparative morphological studies using multiple strains of Glaucocystis were performed until recently (see below).
Although TEM has sufficiently high resolution to elucidate precise characteristics, even in 10-μm-scale microalgae, conventional TEM can reveal only limited parts of cells, locally, using ultrathin samples into which the electron beam are transmitted3,4,5. On the other hand, scanning electron microscopy (SEM) can reveal the characteristics of the entire cell surface, globally, but conventional SEM does not have sufficiently high resolution to observe ultrastructures in detail10,11. Recently, two types of new-generation EM, ultra-high-resolution (UHR) field-emission (FE)-SEM and ultra-high-voltage electron microscopy (UHVEM), have introduced a new paradigm in the field of biology10,11,12,13,14. Despite this, in some recent studies of microalgal and protozoan taxonomy, the utility of molecular approaches continues to be overemphasised15,16,17,18.
UHR FE-SEM enables ultrafine observations of the entire cell surface even at low accelerating voltages (LV); it also allows in situ surface ultrastructures in numerous cells to be observed all at once10,11,12. Our recent study using LV FE-SEM unveiled species diversity within the flagellate glaucophyte genus Cyanophora, identifying three new species10. However, LV FE-SEM cannot be applied to examine a protoplast enclosed by a cell wall, as in the coccoid glaucophyte genus Glaucocystis19,20. For this, UHVEM, which enables in situ 3D ultrastructural observation by thick-section tomography using an ultra-high accelerating voltage, can be used5. Recently, 3D UHVEM tomography revealed morphological diversity in terms of the 3D ultrastructure of the protoplast periphery using three divergent strains of Glaucocystis13,14. Thus, undescribed species of this genus are expected to be delineated morphologically among strains distributed across the globe, based on new-generation EM characteristics.
Here, we aimed to delineate morphologically and phylogenetically different Glaucocystis species based on the combination of several types of microscopy, including 3D UHVEM tomography and LV FE-SEM, combined with molecular phylogenetic results, from 10 globally distributed strains labelled G. nostochinearum Itzigs. ex Rabenh. (1866)21,22 and three newly established strains of Glaucocystis (Supplementary Table 1). A new taxonomic system of Glaucocystis species delineated using new-generation EM is described in this report (Table 1).
Results
Light microscopy
Using LM on the 13 strains (Supplementary Table 1), two Glaucocystis species were identified based on the traditional taxonomic system19,23,24 (Supplementary Table 2): G. nostochinearum and G. oocystiformis Prescott (1944)23 (see Supplementary Note). Moreover, we found differences that could contribute to species delineation within G. nostochinearum in our new taxonomic system (Table 1; Fig. 1; Supplementary Figs 1,2; Supplementary Note).
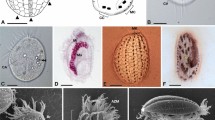
Shown at the same magnification. Scale bar, 20 μm. Note that each colony is enclosed by mother cell wall (arrows) tightly (c,f) or arranged in a less crowded way within an extended mother cell wall (arrowheads) (a,b,d,e). (a) G. geitleri E.G.Pringsh. ex Tos.Takah. & Nozaki sp. nov. strain SAG 229-1. (b) G. nostochinearum Itzigs. ex Rabenh. strain SAG 16.98. (c) G. incrassata (Lemmerm.) Tos.Takah. & Nozaki stat. nov. strain SAG 229-2. (d) G. oocystiformis Prescott strain 126. (e) G. miyajii Tos.Takah. & Nozaki sp. nov. strain Thu10. (f) G. bhattacharyae Tos.Takah. & Nozaki sp. nov. strain 118.
Field-emission scanning electron microscopy
The cell wall of Glaucocystis is composed of cellulose filaments and has the highest cellulose Iα crystallite content of all organisms25,26,27,28. The cellulose filament structure derived from this alga was previously examined by TEM and several types of spectroscopy25,26,27,28,29,30. However, FE-SEM was not yet used to reveal the in situ ultrastructural surface of the Glaucocystis colony or mother cell wall.
Using LV FE-SEM, we detected cellulose filaments of the mother cell wall on the surface of Glaucocystis colonies (Fig. 2). The fibrils on the colony surface were essentially identical in shape among the strains examined, but two types of filament arrangements were recognised. The entire colony surface generally exhibited a gauze fabric-like appearance, with small spaces between fibrils, in G. geitleri E.G.Pringsh. ex Tos.Takah. & Nozaki sp. nov., G. nostochinearum, G. oocystiformis and G. miyajii Tos.Takah. & Nozaki sp. nov. (Fig. 2a,b,d,e). On the other hand, the fibrils were tightly arranged, with no spaces between them, over nearly the entire colony surface in G. incrassata (Lemmerm.) Tos.Takah. & Nozaki stat. nov. and G. bhattacharyae Tos.Takah. & Nozaki sp. nov. (Fig. 2c,f).
Insets show higher magnification image of the mother cell wall surface (boxed area) at the same magnification. Scale bar, 10 μm and 2 μm (insets). Note that each colony is enclosed by a mother cell wall, showing gauze fabric-like fibrils globally (a,b,d,e) or tightly arranged fibrils (c,f ). (a) G. geitleri E.G.Pringsh. ex Tos.Takah. sp. nov. strain SAG 229-1. (b) G. nostochinearum Itzigs. ex Rabenh. strain SAG 16.98. (c) G. incrassata (Lemmerm.) Tos.Takah. stat. nov. strain SAG 229-2. (d) G. oocystiformis Prescott strain 126. (e) G. miyajii Tos.Takah. sp. nov. strain Thu10. (f ) G. bhattacharyae Tos.Takah. sp. nov. strain 118.
This ultrastructural difference in the mother cell wall (Fig. 2) is consistent with the difference in expansion of the mother cell wall observed under LM (Fig. 1; Table 1; Supplementary Note).
Ultra-high-voltage electron microscopy and ultrathin-section transmission electron microscopy
Recent reports13,14 using UHVEM tomography clearly revealed the 3D ultrastructural features of the plasma membrane and the flattened vesicles at the protoplast periphery in three strains or species of Glaucocystis (G. geitleri strain SAG 229-1, G. nostochinearum strain SAG 16.98 and G. incrassata strain SAG 229-2), even though the protoplast was tightly enclosed by a cell wall. In addition, the 3D ultrastructures of the protoplast periphery in these three strains are diverse and can be classified into three types (periphery types A, B and C)13,14. Since these three strains were found to represent three different species (G. geitleri strain SAG 229-1 of type A, G. nostochinearum strain SAG 16.98 of type B and G. incrassata strain SAG 229-2 of type C), they were assigned as authentic strains for these species (see below).
To examine the peripheral 3D ultrastructure of protoplasts in the other three Glaucocystis species, we observed various regions of mature vegetative cells in three strains representing the three species (G. oocystiformis strain 126, G. miyajii strain Thu10 and G. bhattacharyae strain 118, designated here as the authentic strains for the three species; see below) by UHVEM and tomography, as well as ultrathin section TEM (Table 1; Fig. 3; Supplementary Videos 1–3; Supplementary Fig. 3). The protoplast periphery of these species was similar to that in the former three species examined previously. The flattened vesicles were leaflet-like in shape, lacked a plate-like interior structure, and were distributed throughout the entire protoplast periphery just underneath the single-layered plasma membrane (except for the region near basal bodies), but they did not completely enclose the protoplast periphery to form small spaces between the vesicles at the protoplast periphery. In addition, based upon the present UHVEM tomography, G. oocystiformis strain 126 was assigned to periphery type A, whereas G. miyajii strain Thu10 and G. bhattacharyae strain 118 were assigned to periphery type C. Based on the native 3D ultrastructural features of the protoplast periphery established by previous and present studies using UHVEM tomography13,14, the three periphery types are evident and distinguishable from each other, even based on ultrathin-section TEM alone. Thus, peripheral protoplast types were determined in other strains based on ultrathin-section TEM alone; each of the six species exhibited only a single periphery type, despite being composed of more than one strain. G. nostochinearum (periphery type B) and G. miyajii (periphery type C) were clearly distinguished from each other based on the difference in the periphery type, although they were indistinguishable under LM alone (Table 1; Figs 1 and 4; Supplementary Fig. 1).
Electron tomography of protoplast periphery of vegetative cells of G. oocystiformis strain 126 (a,b), G. miyajii Tos.Takah. & Nozaki sp. nov. strain Thu10 (c,d) and G. bhattacharyae Tos.Takah. & Nozaki sp. nov. strain 118 (e,f). Note that G. oocystiformis exhibits periphery type A whereas G. miyajii and G. bhattacharyae exhibit periphery type C (Supplementary Fig. 4). See also Supplementary Videos 1–3. (a,c,e) Ultra-high voltage electron microscopic images. Insets show higher magnification image of the cell periphery (boxed area). Scale bar, 5 μm and 1 μm (insets). (b,d,f ) Tomographic images of boxed area in (a,c,e), respectively. Shown at the same magnification. Scale bar, 1 μm. M, mitochondrion; P, plastid; V, vacuole; W, cell wall. Arrows indicate bar-like grooves of plasma membrane covered by invaginations of flattened vesicles.
Six Glaucocystis (G.) species were classified based on morphological characteristics and molecular phylogeny of cultured material. Colony surface (blue) exhibit two cellulose fibril types under FE-SEM (Fig. 2): gauze fabric-like fibrils (left) and tightly arranged fibrils (right), indicated by two background designs. Periphery types A–C are distinguishable under UHVEM and TEM (Supplementary Fig. 4)13,14, indicated by enlarged diagram of flattened vesicles (yellow) in each species. Four types of cell wall shape at the cell poles are recognised under LM (Supplementary Fig. 1). Phylogeny is based on phylogenetic tree of the concatenated gene sequences (Supplementary Fig. 5); each species exhibits sufficient genetic distances from other species (Supplementary Figs 6–8).
Molecular phylogenetic analyses
The phylogenetic tree of the concatenated plastid gene sequences (Supplementary Fig. 5) demonstrated that 13 Glaucocystis strains could be subdivided into six phylogenetic groups [four robust monophyletic groups and two independent operational taxonomic units (OTUs)], which are essentially equivalent to the G1–G6 groups recognised previously31. These six groups corresponded to the six species delineated by our comparative morphological analysis (Table 1).
In the phylogenetic tree, basal phylogenetic relationships were robustly resolved (with bootstrap values of 82–100%; Supplementary Fig. 5); G. geitleri and G. incrassata occupied the most and second most basal positions, respectively, whereas the other four species (G. nostochinearum, G. oocystiformis, G. miyajii and G. bhattacharyae) represent a large robust monophyletic group (crown lineage), supported by bootstrap values of 100% in neighbour-joining (NJ) and maximum likelihood (ML) analyses. However, the phylogenetic relationships of the four species were not well resolved within the crown lineage.
Internal transcribed spacer-2 secondary structure and genetic distances
The six species of Glaucocystis were evaluated by compensatory base changes (CBCs) in the secondary structure of the internal transcribed spacer (ITS)-2 of nuclear ribosomal DNA (rDNA) (Supplementary Figs 6 and 7) and the genetic distances of a plastid gene (Supplementary Fig. 8). Four Glaucocystis species within the crown lineage exhibited CBCs and sufficient genetic distances to be classified as four distinct species (Supplementary Note).
Discussion
Based on the present comparative morphological and molecular examinations of cultured materials from the genus Glaucocystis, six species were clearly delineated (Table 1; Fig. 4). In contrast to previous reports8,9, that 45 to 50 years ago had to rely on conventional EM only, ultrastructural diversity of the protoplast periphery was significant within the genus Glaucocystis when examined by UHVEM (Figs 3 and 4; Supplementary Figs 3 and 4). Moreover, ultrastructural diversity was clarified in the arrangement of cellulose filaments of the mother cell under LV FE-SEM (Figs 1 and 2). Based on the differences in these new-generation EM characteristics and LM features of the 13 Glaucocystis strains, we could delineate six morphological species that correspond to six phylogenetic groups (G1–G6) recognised by previous31 and present phylogenetic analyses (Table 1; Fig. 4; Supplementary Fig. 5). Although G. oocystiformis can be easily distinguished from the other five species based on differences in LM characteristics and phylogenetic positions (Table 1; Fig. 1; Supplementary Figs 1 and 5), the in situ ultrastructural features of G. oocystiformis are similar to those of G. geitleri in having periphery type A and gauze fabric-like fibrils (Supplementary Fig. 5). Among the other four species, G. bhattacharyae and G. incrassata have essentially the same in situ ultrastructures (periphery type C and tightly arranged fibrils), although they were distinguished from one another based on differences in LM characteristics (Table 1; Supplementary Fig. 1). Thus, in addition to new-generation EM observations, LM data and molecular phylogenetic analyses are essential for delineating microalgal species.
The novel strains established here from a single field sample were classified into three species (Table 1; Supplementary Table 1; Supplementary Fig. 5). Although G. nostochinearum has been considered a cosmopolitan species24,32,33, the records may be based on several species, which can be distinguished using the new-generation taxonomic methodology established here (Supplementary Note).
The plasma membrane of five of the six Glaucocystis species had numerous grooves throughout the protoplast surface (Figs 3 and 4; Supplementary Figs 3 and 4)13,14. In G. nostochinearum13, however, the plasma membrane lacked grooves or invaginations (type B; Supplementary Fig. 4) as in Cyanophora species10,11. Since G. nostochinearum belongs to the crown lineage within Glaucocystis (Supplementary Fig. 5), the lack of grooves or invaginations in G. nostochinearum might have evolved secondarily within Glaucocystis (Fig. 4). Vegetative cells of Cyanophora species are apparently smaller than those of Glaucocystis10, and G. nostochinearum exhibits one of the smallest cell sizes within Glaucocystis (Table 1; Fig. 1; Supplementary Fig. 1). In addition, the surface-area-to-volume ratio is smaller (inversely proportional to the cell size) and the transportation of substances across the plasma membrane more limited in larger cells. Thus, the presence of grooves or invaginations at the protoplast periphery in the five Glaucocystis species might contribute to expansion of the surface area of the protoplast and consequently to their large cell size.
The Gloeochaetales are another order of glaucophytes that are characterised by having palmelloid immotile vegetative cells and include two genera, Gloeochaete and Cyanoptyche20; some species have zoospores34,35,36. These algae might represent the evolutionarily intermediate stage between the flagellate Cyanophora (Cyanophorales) and the coccoid Glaucocystis (Glaucocystales). Within the Gloeochaetales, cultured strains labelled Cyanoptyche gloeocystis Pascher37 and Gloeochaete wittrockiana Lagerh38 are available, but do not number more than three in each species ( http://www.ccac.uni-koeln.de/; http://sagdb.uni-goettingen.de/). Although no taxonomic studies have been performed based on EM and/or molecular data, these two taxa are considered cosmopolitan species34,39. Therefore, taxonomic studies based on molecular methods and comparative in situ ultrastructural characteristics using various clonal strains, as in Cyanophora10 and Glaucocystis evaluated here, would be useful for these two species or genera. 3D UHVEM tomography will reveal the peripheral in situ ultrastructures of their vegetative cells even when enclosed by a non-cellulosic extracellular matrix35,36, as in Glaucocystis13,14. LV FE-SEM may be applicable in easily inducible, naked zoospores of the Gloeochaetales35,36 for in situ ultrastructural observation of the protoplast surface, as in Cyanophora10. These two types of new-generation EM should be capable of revealing the actual diversity in ultrastructures in the peripheral protoplast in situ, leading to the delineation of more natural species of the gloeochaetalean algae, when combined with molecular phylogenetic results.
Conclusions
In recent taxonomic work on certain microorganisms, there has been a tendency to avoid morphological approaches in favour of molecular ones15,16,17,18. However, species delineation based only on molecular data cannot demonstrate how the species live. Even when whole-genome sequence data are available, we can only speculate on the metabolic pathways employed by the organism. Even in bacteria/archaea, species delineation has been carried out on the basis of phenotypic characteristics40,41. Next-generation microbial taxonomy, which is just now becoming established, utilises new-generation EM methods (e.g. FE-SEM and UHVEM) to demonstrate detailed in situ ultrastructural features of microscopic organisms in their entirety. Molecular barcoding is only meaningful for lineages within which species have already been delineated and recognised by morphological or phenotypic characteristics. Global and in situ ultrafine microscopy should become the mainstream method used to delineate microbial species, as in the present study on Glaucocystis.
Taxonomic Accounts
Glaucocystis nostochinearum Itzigs. ex Rabenh. (in Alg. Eur. 94–5: no. 1935. 1866)21,22.
Syntypes: Rabenhorst’s exsiccata, Die Algen Europas21 packet no. 1935.
Lectotype (here designated): the permanent slide R1935J prepared from a syntype of Farlow Herbarium, University of Harvard (FH), deposited in FH (Supplementary Fig. 2; Supplementary Note).
Syntypic authentic strain: not available.
Type locality: Berlin, Prussia (now, Germany).
Epitype (here designated): Resin-embedded cells of the new authentic strain SAG 16.98, deposited as TNS-AL-58925 in Department of Botany, National Museum of Nature and Science (TNS).
Epitypic authentic strain (here designated): SAG 16.98.
Epitype locality: Lower Saxony, pond in quarry at Walkenried/Harz, surface of Myriophyllum sp., Germany.
Glaucocystis oocystiformis Prescott (in Farlowia 1(3): 372. 1944)23.
Holotype: Prescott 1944. Farlowia. pl. 4, Fig. 20.
Holotypic authentic strain: not available.
Type locality: Trout Lake, Vilas County, Wisconsin, USA.
Epitype (here designated): Resin-embedded cells of the new authentic strain 126, deposited as TNS-AL-58926 in TNS.
Epitypic authentic strain (here designated): Isolate 126, also available as NIES-3868 (Supplementary Table 1).
Epitype locality: Funabashi-shi, Chiba, Japan (35.694283°N, 140.048166°E).
Glaucocystis incrassata (Lemmerm.) Tos.Takah. & Nozaki stat. nov.
Basionym: Glaucocystis nostochinearum var. incrassata Lemmerm. (in Arch. Hydrobiol. Planktonkd. 4: 178. 1908)42.
Holotype: Lemmermann, Arch. Hydrobiol. Planktonkd. 4: 178. 1908, Taf. V., Fig. 442.
Holotypic authentic strain: not available.
Type locality: Lentini, Sicily, Italy.
Epitype (here designated): Resin-embedded cells of the new authentic strain SAG 229-2, deposited as TNS-AL-58923 in TNS.
Epitypic authentic strain ( here designated): SAG 229-2.
Epitype locality: Denmark.
Glaucocystis geitleri E.G.Pringsh. ex Tos.Takah. & Nozaki sp. nov.
≡ Glaucocystis geitleri E.G.Pringsh. (in Stud. Pl. Physiol. 1958)43 nom. provis., inval. (see Supplementary Note).
Diagnosis:
Coccoid alga, enclosed by cellulosic cell wall; solitary or colonial generally with two cells. Cells ca. 30–40 μm long × 20–30 μm wide, truncate-ellipsoidal, often with clear polar thickenings, lacking polar nodules and equatorial ring. Two vestigial flagella between cell wall and protoplast periphery, positioned at equator of cells. Protoplast periphery, with numerous small depressions arranged regularly. Depression at intervals of ca. 500–800 nm, shared by plasma membrane and centre of underlying flattened vesicle. Flattened vesicles leaflet-like, not overlapping one another. Colony lacking attaching stalk; mother cell wall extended prominently, with a gauze fabric-like appearance and small spaces between fibrils.
Type locality: Cambridge, England, UK.
Holotype: Resin-embedded cells of the new authentic strain SAG 229-1, deposited as TNS-AL-58922 in TNS.
Holotypic authentic strain: SAG 229-1.
Glaucocystis miyajii Tos.Takah. & Nozaki sp. nov.
Diagnosis:
Coccoid alga, enclosed by cellulosic cell wall; solitary or colonial generally with four cells. Cells ca. 19–24 μm long × 10–15 μm wide, ellipsoidal, lacking polar thickenings, polar nodules and equatorial ring. Two vestigial flagella between cell wall and protoplast periphery, positioned at equator of cells. Protoplast periphery, with numerous small depressions arranged regularly. Depression at intervals of ca. 200–600 nm, shared by plasma membrane and centre of underlying flattened vesicle. Flattened vesicles leaflet-like, slightly overlapping one another. Colony lacking attaching stalk; mother cell wall extended prominently, with a gauze fabric-like appearance and small spaces between fibrils.
Type locality: Funabashi-shi, Chiba, Japan (35.694283°N, 140.048166°E).
Holotype: Resin-embedded cells of the new authentic strain Thu10, deposited as TNS-AL-58924 in TNS.
Holotypic authentic strain: Isolate Thu10, also available as NIES-3867 (Supplementary Table 1).
Etymology: Named after Prof. Kazuyuki Miyaji (University of Toho), who contributed much to phycology.
Glaucocystis bhattacharyae Tos.Takah. & Nozaki sp. nov.
Diagnosis:
Coccoid alga, enclosed by cellulosic cell wall; solitary or colonial generally with four cells. Cells ca. 17–27 μm long × 12–22 μm wide, truncate-ellipsoidal, lacking polar thickenings, polar nodules and equatorial ring. Two vestigial flagella between cell wall and protoplast periphery, positioned at equator of cells. Protoplast periphery, with numerous small depressions arranged regularly. Depression at intervals of ca. 200–600 nm, shared by plasma membrane and centre of underlying flattened vesicle. Flattened vesicles leaflet-like, slightly overlapping one another. Colony lacking attaching stalk; mother cell wall lacking prominent extension and a gauze fabric-like appearance, with tightly arranged fibrils and no spaces between fibrils.
Type locality: Funabashi-shi, Chiba, Japan (35.694283°N, 140.048166°E).
Holotype: Resin-embedded cells of the new authentic strain 118, deposited as TNS-AL-58921 in TNS.
Holotypic authentic strain: Isolate 118, also available as NIES-3866 (Supplementary Table 1).
Etymology: Named after Prof. Debashish Bhattacharya (Rutgers University), who contributed much to phycology.
Key to species of Glaucocystis
Based on Table 1 and Supplementary Table 2.
A. Colony with stalk-----------------------------------------------------------------------------B.
A. Colony without stalk--------------------------------------------------------------------------C.
B. Cell shape ellipsoidal-----------------------------------------------------G. indica R.J.Patel
B. Cell shape kidney-shaped--------------------------------------------------------------------------------------------------------------G. reniformis B.N.Prasad, R.K.Mehrotra & P.K.Misra
C. Cell wall with equatorial ring-------------------------------------------G. cingulata Bohlin
C. Cell wall without equatorial ring------------------------------------------------------------D.
D. Cell spherical--------------------------------------------------------------G. duplex Prescott
D. Cell ellipsoidal---------------------------------------------------------------------------------E.
E. Cell measured 10–18 × 6–10 μm-------------------------------------------G. bullosa Wille
E. Cell measured 17–50 × 10–30 μm----------------------------------------------------------F.
F. Cell wall with polar nodules--------------------------------------G. oocystiformis Prescott
F. Cell wall without polar nodules--------------------------------------------------------------G.
G. Cell wall with polar thickenings------------------------------------------------------------H.
G. Cell wall without polar thickenings----------------------------------------------------------I.
H. Mother cell wall with prominent extension, having a gauze fabric-like appearance and small spaces between fibrils; cell measured 30–50 × 19–30 μm; grooves at intervals of 500–800 nm; vesicles not overlapping---------------------------------------------------------------------------------------------G. geitleri E.G.Pringsh. ex Tos.Takah. & Nozaki sp. nov.
H. Mother cell wall without prominent extension, lacking a gauze fabric-like appearance and spaces between fibrils; cell measured 22–32 × 15–24 μm; grooves at intervals of 200–600 nm; vesicles frequently overlapping-----------------------------------------------------------------------G. incrassata (Lemmerm.) Tos.Takah. & Nozaki stat. nov.
I. Poles of cell truncate; mother cell wall without prominent extension, lacking a gauze fabric-like appearance and spaces between fibrils---------------------------------------------------------------------------------------------G. bhattacharyae Tos.Takah. & Nozaki sp. nov.
I. Poles of cell not truncate; mother cell wall with prominent extension, having a gauze fabric-like appearance and small spaces between fibrils-------------------------------------J.
J. Protoplast periphery with grooves-------------G. miyajii Tos.Takah. & Nozaki sp. nov.
J. Protoplast periphery without grooves------------G. nostochinearum Itzigs. ex Rabenh.
Methods
Strains and culture conditions for observation
Ten culture strains of Glaucocystis were obtained from public culture collections (Supplementary Table 1) at the National Institute for Environmental Studies (NIES, http://mcc.nies.go.jp/)44 and the Sammlung von Algenkulturen der Universität Göttingen (SAG, http://sagdb.uni-goettingen.de/)45,46. We also used three strains of Glaucocystis newly established from freshwater samples collected in Japan (strains 118, 126 and Thu10; Supplementary Table 1). The cultures were maintained as described previously13.
Light microscopy
Permanent slides were prepared using air-dried cells from the syntype material of G. nostochinearum and cultured material from authentic strains. Rehydrated cells from the syntype material or cultured cells were fixed with 2% glutaraldehyde in medium and washed with medium and distilled water on 0.1%-poly-L-lysine-coated 18-mm micro-cover glasses (Matsunami Glass Ind., Ltd., Kishiwada, Japan). After dehydration using a graded ethanol series and infiltration with xylene, the glass was covered with 60 °C Canada balsam xylene, placed on the 60 °C Canada balsam on a glass slide, and then the slide was incubated at 60 °C for a few days. LM observations were carried out as described previously10 using the permanent slides and living cultured cells.
Field-emission scanning electron microscopy
LV FE-SEM was performed as described previously11 using all 13 Glaucocystis strains, but cells were harvested directly, treated with the critical point dryer JCPD-5 (JEOL) and observed using the UHR FE-SEM SU8220 (Hitachi High-Technologies, Tokyo, Japan).
Transmission electron microscopy and ultra-high-voltage electron microscopy
Since the high-pressure freezing and freeze-substitution fixation method is generally expected to be superior to chemical fixation in preserving the integrity of cellular ultrastructures12, this method was performed for TEM and UHVEM as described previously13. Ultrathin-section TEM was also performed as described previously13 for all 13 Glaucocystis strains. In addition, UHVEM and reconstruction of the tomographic images were carried out as described previously13 in three authentic strains of three Glaucocystis species (Fig. 3).
Molecular phylogenetic analysis and comparative analysis of the secondary structure of ITS-2 in nuclear ribosomal DNA
DNA extraction, polymerase chain reaction (PCR) and direct sequencing of the PCR products were performed as described previously10,47,48, using primers designed in a previous study10,31,49. The secondary structure of nuclear rDNA ITS-2 was constructed as described previously10. Phylogenetic relationships between Glaucocystis species were examined based on analyses of the concatenated sequences (2,211 base pairs) of the photosystem I P700 chlorophyll a apoprotein A2 (psaB) gene (1,461 base pairs) and the photosystem II P680 chlorophyll a apoprotein D1 (psbA) gene (750 base pairs) from 13 strains of Glaucocystis, representing 10 OTUs (based on identical sequences), and three strains of three other glaucophyte genera as an outgroup (Supplementary Table 1). The sequences were aligned as described previously10 and subjected to phylogenetic analyses. ML and NJ analyses were performed as described previously10, except that one selected model was used: the general time reversible + gamma model with invariant sites for ML.
Additional Information
How to cite this article: Takahashi, T. et al. Delineation of six species of the primitive algal genus Glaucocystis based on in situ ultrastructural characteristics. Sci. Rep. 6, 29209; doi: 10.1038/srep29209 (2016).
References
Van Leeuwenhoeck, A. Observations, communicated to the publisher by Mr. Antony van Leeuwenhoeck, in a Dutch letter of the 9th of Octob. 1676. Here English’d: concerning little animals by him observed in rain-well-sea- and snow- water; as also in water wherein pepper had lain infused. Phil. Trans. 12, 821–831 (1677).
Smit, P. & Heniger, J. Antoni van Leeuwenhoek (1632–1723) and the discovery of bacteria. Antonie Van Leeuwenhoek 41, 217–228 (1975).
Alberts, B. et al. Molecular Biology of the Cell, 4th Edition (Garland Science, New York, 2002).
Mulvey, T. Advances in Imaging and Electron Physics. the Growth of Electron Microscopy. Volume 96 (Academic Press, London, 1996).
Cyranoski, D. Microscopic marvels: The big and the bold. Nature 459, 634–635 (2009).
Adl, S. M. et al. Diversity, nomenclature, and taxonomy of protists. Sys. Biol. 56, 684–689 (2007).
Adl, S. M. et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–514 (2012).
Schnepf, E., Koch, W. & Deichgräber, G. Zur Cytologie und taxonomischen Einordnung von Glaucocystis . Arch. Microbiol. 55, 149–174 (1966).
Schnepf, E. & Brown Jr., R. M. In Origin and continuity of cell organelles (eds Reinert, J. & Ursprung, H. ) 299–322 (Springer-Verlag, Berlin, 1971).
Takahashi, T. et al. Five Cyanophora (Cyanophorales, Glaucophyta) species delineated based on morphological and molecular data. J. Phycol. 50, 1058–1069 (2014).
Takahashi, T., Sato, M., Toyooka, K. & Nozaki, H. Surface ornamentation of Cyanophora paradoxa (Cyanophorales, Glaucophyta) cells as revealed by ultra-high resolution field emission scanning electron microscopy. Cytologia 79, 119–123 (2014).
Osumi, M., Konomi, M., Sugawara, T., Takagi, T. & Baba, M. High-pressure freezing is a powerful tool for visualization of Schizosaccharomyces pombe cells: ultra-low temperature and low-voltage scanning electron microscopy and immunoelectron microscopy. J. Electron Microsc. 55, 75–88 (2006).
Takahashi, T., Nishida, T., Saito, C., Yasuda, H. & Nozaki, H. Ultra-high voltage electron microscopy of primitive algae illuminates 3D ultrastructures of the first photosynthetic eukaryote. Sci. Rep. 5, 14735 (2015).
Takahashi, T., Nishida, T., Saito, C., Yasuda, H. & Nozaki, H. A new type of 3D peripheral ultrastructure in Glaucocystis (Glaucocystales, Glaucophyta) as revealed by ultra-high voltage electron microscopy. J. Phycol. 52, 486–490 (2016).
Smith, M. A. et al. Barcoding a quantified food web: crypsis, concepts, ecology and hypotheses. PLoS One 6, e14424 (2011).
Wełnicz, W., Grohme, M. A., Kaczmarek, Ł., Schill, R. O. & Frohme, M. ITS-2 and 18S rRNA data from Macrobiotus polonicus and Milnesium tardigradum (Eutardigrada, Tardigrada). J. Zool. Syst. Evol. Res. 49, 34–39 (2011).
Schill, R. O., Förster, F., Dandekar, T. & Wolf, M. Using compensatory base change analysis of internal transcribed spacer 2 secondary structures to identify three new species in Paramacrobiotus (Tardigrada). Organisms Divers. Evol. 10, 287–296 (2010).
Rybalka, N., Wolf, M., Andersen, R. A. & Friedl, T. Congruence of chloroplast- and nuclear-encoded DNA sequence variations used to assess species boundaries in the soil microalga Heterococcus (Stramenopiles, Xanthophyceae). BMC Evol. Biol. 13, 39 (2013).
Komárek, J. & Fott, B. Chlorophyceae (Grünalgen); Ordnung: Chlorococcales 446–555 (E. Schweizerbart’sche Verlagsbuchhandlung, Stuttgart, 1983).
Kies, L. & Kremer, B. P. Typification of the Glaucocystophyta. Taxon 35, 128–133 (1986).
Rabenhorst, L. G. Die Algen Europas, Fortsetzung der Algen Sachsens, resp. Mittel-europas. Decades 94–95, Number 1935 (Dresden, 1866).
Rabenhorst, L. G. Flora Europaea Algarum Aquae Dulcis et Submarinae. III. Algas Chlorophyllophyceas, Melanophyceas et Rhodophyceas Complectens (Eduard Kummer, Leipzig, 1868).
Prescott, G. W. New species and varieties of Wisconsin algae. Farlowia 1, 347–385 (1944).
Prescott, G. W. Algae of the Western Great Lakes Area (WC Brown Company, Dubuque, 1962).
Sugiyama, J., Persson, J. & Chanzy, H. Combined infrared and electron diffraction study of the polymorphism of native celluloses. Macromolecules 24, 2461–2466 (1991).
Imai, T., Sugiyama, J., Itoh, T. & Horii, F. Almost Pure Iα Cellulose in the Cell Wall of Glaucocystis . J. Struct. Biol. 127, 248–257 (1999).
Briois, B. et al. Iα → Iβ transition of cellulose under ultrasonic radiation. Cellulose 20, 597–603 (2013).
Lee, C. M., Kafle, K., Park, Y. B. & Kim, S. H. Probing crystal structure and mesoscale assembly of cellulose microfibrils in plant cell walls, tunicate tests, and bacterial films using vibrational Sum Frequency Generation (SFG) spectroscopy. Phys. Chem. Chem. Phys. 16, 10844–10853 (2014).
Schnepf, E. Struktur der Zellwände und Cellulosefibrillen bei Glaucocystis . Planta 67, 213–224 (1965).
Willison, J. H. M. & Brown Jr., R. M. Cell wall structure and deposition in Glaucocystis . J. Cell Biol. 77, 103–119 (1978).
Chong, J., Jackson, C., Kim, J. I., Yoon, H. S. & Reyes-Prieto, A. Molecular markers from different genomic compartments reveal cryptic diversity within glaucophyte species. Mol. Phylogenet. Evol. 76, 181–188 (2014).
Prasad, B. N. Glaucocystis nostochinearum (Itzig.) Rabenhorst in India. Bull. Bot. Soc. 13, 44–45 (1961).
Starmach, K. Cyanophyta - Sinice. Glaucophyta - Glaukofity. (Państwowe Wydawn. Naukowe, Warszawa, 1966).
Lagerheim, G. Gloeochaete Lagerheim und Schrammia Dangeard. Nouva Notarisia 1, 227–231 (1890).
Kies, L. Zur systematischen Einordnung von Cyanophora paradoxa, Gloeochaete wittrockiana und Glaucocystis nostochinearum. Ber. Dtsch. Bot. Ges. 92, 445–454 (1979).
Kies, L. Ultrastructure of Cyanoptyche gloeocystis f. dispersa (Glaucocystophyceae). Plant Syst. Evol. 164, 65–73 (1989).
Pascher, A. Über einige Endosymbiosen von Blaualgen in Einzellern. Jahrb. Wiss. Bot. 71, 386–462 (1929).
Lagerheim, G. Bidrag till Sveriges algflora. Öfvers. Kgl. Svensk. Vetensk 2, 37–78 (1883).
Bourrelly, P. Algues d’eau douce de la République de Côte d’Ivoire. Bull. Inst. Franç. Afr. Noire, sér. A 23, 283–374 (1961).
Honda, T., Fujita, T. & Tonouchi, A. Aminivibrio pyruvatiphilus gen. nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int. J. Syst. Evol. Microbiol. 63, 3679–3686 (2013).
Sizova, M. V. et al. Stomatobaculum longum gen. nov., sp. nov., an obligately anaerobic bacterium from the human oral cavity. Int. J. Syst. Evol. Microbiol. 63, 1450–1456 (2013).
Lemmermann, E. Algologische Beiträge. Arch. Hydrobiol. Planktonkde. 4, 165–192 (1908).
Pringsheim, E. G. In Studies in Plant Physiology (ed. Prat, S. ) 165–184 (Czechoslovak Acad. Sci., Prague, 1958).
Kasai, F. et al. NIES-Collection. List of Strains 8th Edition (Japanese Journal of Phycology, Tsukuba, 2009).
Koch, W. Verzeichnis der Sammlung von Algenkulturen am Pflanzenphysiologischen Institut der Universität Göttingen. Arch. Microbiol. 47, 402–432 (1964).
Schlösser, U. G. SAG – Sammlung von Algenkulturen at the University of Göttingen. Catalogue of Strains 1994. Bot. Acta 107, 113–186 (1994).
Nakada, T. & Nozaki, H. Re-evaluation of three Chlorogonium (Volvocales, Chlorophyceae) species based on 18S ribosomal RNA gene phylogeny. Eur. J. Phycol. 42, 177–182 (2007).
Hayama, M., Nakada, T., Hamaji, T. & Nozaki, H. Morphology, molecular phylogeny and taxonomy of Gonium maiaprilis sp. nov. (Goniaceae, Chlorophyta) from Japan. Phycologia 49, 221–234 (2010).
Nozaki, H. et al. Origin and Evolution of the Colonial Volvocales (Chlorophyceae) as Inferred from Multiple, Chloroplast Gene Sequences. Mol. Phylogenet. Evol. 17, 256–268 (2000).
Acknowledgements
We sincerely appreciate Prof. Kazuyuki Miyaji (Toho University) offering information and helping for collecting algae kindly. We would like to thank Ms. Genevieve E. Tocci, Dr. Robert K. Edgar (Farlow Herbarium, University of Harvard, FH) and Prof. Jin Murata (Herbarium, University of Tokyo, TI) for the loan arrangement and photography permission of G. nostochinearum syntype material. This work was supported partly by “Nanotechnology Network Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan” at the Research Center for Ultrahigh Voltage Electron Microscopy, Osaka University (Handai multi-functional Nano-Foundry), by Grants-in-Aid from the NC-CARP project of MEXT (to CS), and by Grants-in-Aid for Scientific Research on Innovative Areas (number 26117708 to HN) and Scientific Research (A) (number 24247042 to HN) from MEXT/JSPS KAKENHI. We thank Ms. Fumiko Ishitsuna (University of Tokyo) for her kind help for using the high-pressure freezing machine HPM010 and Ms. Ayako Watanabe (University of Tokyo) for her kind guidance of critical point dryer JCPD-5.
Author information
Authors and Affiliations
Contributions
T.T. designed the study and H.N. supervised the project. T.T., C.S. and T.N. prepared the UHVEM and TEM samples. The UHVEM observation and data analysis were performed by T.T. and T.N. and supervised by H.Y. The permanent slides were prepared by T.T. and A.T. The FE-SEM samples were observed by T.T. and M.S., supervised by K.T. Novel Glaucocystis strains and secondary structures of nuclear rDNA ITS-2 of Glaucocystis strains were established by T.T. and R.M. All the other experiments and data analyses were performed by T.T. The manuscript was written by T.T. and H.N. and modified by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Takahashi, T., Nishida, T., Tuji, A. et al. Delineation of six species of the primitive algal genus Glaucocystis based on in situ ultrastructural characteristics. Sci Rep 6, 29209 (2016). https://doi.org/10.1038/srep29209
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep29209
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.