Abstract
Carboxysomes are bacterial microcompartments that enhance carbon fixation by concentrating ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and its substrate CO2 within a proteinaceous shell. They are found in all cyanobacteria, some purple photoautotrophs and many chemoautotrophic bacteria. Carboxysomes consist of a protein shell that encapsulates several hundred molecules of RuBisCO and contain carbonic anhydrase and other accessory proteins. Genes coding for carboxysome shell components and the encapsulated proteins are typically found together in an operon. The α-carboxysome operon is embedded in a cluster of additional, conserved genes that are presumably related to its function. In many chemoautotrophs, products of the expanded carboxysome locus include CbbO and CbbQ, a member of the AAA+ domain superfamily. We bioinformatically identified subtypes of CbbQ proteins and show that their genes frequently co-occur with both Form IA and Form II RuBisCO. The α-carboxysome-associated ortholog, CsoCbbQ, from Halothiobacillus neapolitanus forms a hexamer in solution and hydrolyzes ATP. The crystal structure shows that CsoCbbQ is a hexamer of the typical AAA+ domain; the additional C-terminal domain, diagnostic of the CbbQ subfamily, structurally fills the inter-monomer gaps, resulting in a distinctly hexagonal shape. We show that CsoCbbQ interacts with CsoCbbO and is a component of the carboxysome shell, the first example of ATPase activity associated with a bacterial microcompartment.
Similar content being viewed by others
Introduction
The enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) plays an essential role in the global carbon cycle by catalyzing the fixation of CO2 onto ribulose-1,5-bisphosphate and subsequent conversion of the intermediate to 3-phosphoglycerate (3-PGA) in the first step of the Calvin-Benson-Bassham (CBB) Cycle. This conversion is sufficiently slow to serve as the rate-limiting step of the CBB Cycle and, moreover, RuBisCO, is competitively inhibited by oxygen1,2,3. To compensate for the inefficiency of RuBisCO, cyanobacteria, some purple phototrophs and many chemoautotrophs have evolved a carbon concentrating mechanism that involves transporters to elevate cytosolic inorganic carbon levels and carboxysomes, bacterial microcompartments (BMCs) that sequester RuBisCO within a protein shell4,5,6,7,8. Cytosolic bicarbonate diffuses into the carboxysome where it is converted into CO2 by a resident carbonic anhydrase; this effectively elevates the substrate concentration near the active site of RuBisCO. The proteinaceous shell is thought to limit the loss of CO29 and may restrict access of inhibitory O2.
Two types of carboxysomes have been classified based on the type of RuBisCO they encapsulate: Form IA RuBisCO in α-carboxysomes and Form IB RuBisCO in β-carboxysomes10. The model organism for α-carboxysome research is the sulfur-oxidizing bacterium Halothiobacillus neapolitanus (H. nea) because it is genetically tractable and because H. nea carboxysomes can be purified in high yield. Genes encoding α-carboxysome components constitute an operon, the cso operon11,12, which in H. nea includes the genes for the Form IA RuBisCO large and small subunits (cbbL and cbbS), the large shell protein CsoS2 that is essential for carboxysome assembly (csoS2)13, the shell-associated β-carbonic anhydrase CsoSCA (csoS3)14, the pentameric CsoS4A and CsoS4B proteins that form the vertices of the carboxysome shell (csoS4A and csoS4B)15,16 and the hexameric shell subunit assemblies of CsoS1A, CsoS1B, CsoS1C (csoS1C, csoS1A and csoS1B) (Fig. 1a)12.
Recently structural and biochemical evidence demonstrated the presence of a pseudohexameric shell protein, CsoS1D, in the carboxysomes of the marine cyanobacterium Prochlorococcus marinus MED417,18. This protein is encoded by a gene located upstream of the cso operon in α-cyanobacteria and downstream of the cso operon in H. nea (Fig. 1a), suggesting that it is likely differentially regulated19. Bioinformatic analysis has likewise expanded the locus of potential carboxysome-associated genes beyond the cso operon13,18. One of these, the pterin-4α-carbinolamine dehydratase-like (pcd-like) is absolutely conserved among all α-carboxysome gene clusters18. Its gene product was recently identified as a potential α-carboxysome RuBisCO assembly factor; it improved recombinant RuBisCO solubility when co-expressed with the GroEL/ES chaperonin complex in E. coli20. The need for such assembly factors for proper RuBisCO folding in plants and cyanobacteria is well documented; presumably they are required in chemoautotrophs as well21,22.
Another member of the expanded α-carboxysome locus is CbbQ, a member of the AAA+ superfamily proteins (ATPases Associated with diverse cellular Activities), which are found across all kingdoms of life and fulfill a variety of functions, mostly involved in ATP-driven dissociation, unfolding and remodeling of macromolecules23,24. Members of the superfamily share a structurally conserved ATPase domain of about 250 amino acids, which typically consists of a large N-terminal P-loop NTPase αβα domain and a smaller, all-α-helical, C-terminal domain. There are several key regions involved in ATP binding and hydrolysis which are highly conserved among all members of the family, including Walker A, Walker B and arginine finger motifs. AAA+ proteins typically form cyclic hexamers in the presence of nucleotides and are active in this state. The nucleotide binding pocket is located between two subdomains and also at the interface between neighboring subunits. Hydrolysis and nucleotide release trigger conformational changes underlying the various functions.
One well characterized AAA+ domain associated with bacterial RuBisCO is RuBisCO activase (CbbX). CbbQ and CbbX (from H. nea and Rhodobacter sphaeroides, respectively) are only 18% identical, with the majority of homology in the AAA+ domain. In contrast to CbbX, there is relatively little experimental data available on the function of CbbQ. The CbbQ from Hydrogenophilus thermoluteolus was shown to have ATP binding and hydrolysis activity; it was estimated to be a decamer by size exclusion chromatography25. Orthologs from Hydrogenophilus thermoluteolus (formerly called Pseudomonas hydrogenothermophila) that are associated with noncarboxysomal form I RuBisCO and two paralogs from Hydrogenovibrio marinus strain MH-110, associated with noncarboxysomal form I and form II RuBisCO, respectively, have been characterized. When they were co-expressed in E. coli with RuBisCO, there was a modest increase in RuBisCO activity26,27. Due to the difficulties of expressing functional RuBisCO in E. coli, which typically requires co-expression of chaperones, it is unclear whether this increase in activity would be the same in a native host. To date, the effect of CbbQ on the carboxysomal RuBisCO and on carboxysome function is unknown.
Here we report the characterization of the CbbQ ortholog that is associated with the α-carboxysome locus of H. nea (Hneap_0905). Bioinformatically we show that carboxysome locus-associated CbbQ (CsoCbbQ) orthologs form a distinct clade of CbbQ proteins. H. nea CsoCbbQ forms a complex when recombinantly co-expressed with CbbO, another member of the expanded H. nea α-carboxysome locus. We also determined the crystal structure of the H. nea csocbbQ gene product and confirmed ATPase activity in the recombinant protein. A deletion mutant in H. nea was generated to examine the potential role of CbbQ in carboxysome function. We found that CbbQ is tightly associated with the carboxysome shell, indicating that the structure and possibly the function of the carboxysome shell is more complex than previously thought. Our results suggest that a component of the α-carboxysome shell has ATPase activity.
Results
Bioinformatic Characterization of CbbQ Orthologs
We identified CbbQ orthologs in a variety of autotrophic bacteria that encode Form IA or Form II RuBisCO (Supplementary Fig. 1). The defining features are an N-terminal AAA+ domain (pfam07728) containing the characteristic residues and motifs for ATP binding and a C-terminal domain confined to CbbQ members of the AAA+ superfamily (pfam08406). Phylogenetically, the CbbQ orthologs fall into four classes: those encoded proximal to the genes for 1) non-carboxysomal Form IA RuBisCO, 2) Form II RuBisCO, 3) the α-carboxysome superlocus (csocbbQ). The fourth class consisted of remote homologs not associated with RuBisCO or carboxysomes (Table 1). The H. nea genome encodes a carboxysome-associated and a Form II RuBisCO-associated CbbQ. The primary structures of the two CbbQ paralogs in H. nea are 71% identical. They are, however, less closely related to each other than to their orthologs from comparable genetic contexts (i.e. carboxysomal locus-associated or Form II-associated). The Form II-associated CbbQs form a distinct clade separate from the ones associated with carboxysomal and non-carboxysomal Form I RuBisCO (Supplementary Fig. 1, blue). Several CbbQ homologs in carboxysome-containing organisms are not found in any of those categories and form a separate clade (with the exception of Thimo_0165, which falls into the Form I RuBisCO-associated clade (Supplementary Fig. 1, grey). There is some overlap between non-carboxysomal Form IA-associated CbbQs and those associated with the carboxysome (Supplementary Fig. 1). Collectively these data implicate CbbQ in the biosynthesis, multimeric assembly or activation of the large subunit of RuBisCO.
Recently, representatives of all distinct types of BMC loci encoded in bacterial genomes were described28. These loci frequently encode not only the proteins forming the microcompartment, but also transporters, regulators and other gene products that support the function of the BMC. The α-carboxysome locus of Acidithiobacillus caldus typifies that found in chemoautotrophs (Fig. 1b). The α-carboxysome locus of H. nea is organized slightly differently, but contains all of the expanded α-carboxysome locus components. The csocbbQ gene in H. nea (Hneap_0905) is separated from the terminal gene of the cluster, csoS1D by a gene for a hypothetical protein that is not conserved among loci (Fig. 1a). Between csocbbQ and the canonical carboxysome operon in H. nea there are several genes that are typically conserved in α-carboxysome loci; these include the gene for a pcd-like protein (pfam01329) and genes annotated as homologs of parF/Cbi (pfam01656), cbbO, nuoL (pfam00361), DUF2309 (pfam10070) and NPII/SbtB (pfam00543). CbbO genes are almost always encoded with csocbbQ (Supplementary Table 1, the only exception being A39ODRAFT_01696 from Lamprocystis purpurea DSM 4197), independent of their proximity to other proteins like RuBisCO, suggesting their functions are most likely connected28. Further support for a possible functional interaction between CbbQ and CbbO comes from the observation that von Willebrand Factor A (VWA) domain-containing proteins (such as CbbO) are frequently interaction partners with MoxR-like AAA+ domains, which include CbbQ29,30.
X-ray Crystal Structure Determination
Hexa-histidine-tagged H. nea CsoCbbQ (Hneap_0905) was purified by affinity chromatography and crystallized with ATP in the mother liquor by vapor diffusion in sitting drops. Native data diffracting to 2.8 Å and belonging to space group H3 were collected (Table 2). Despite the presumed high structural homology of the AAA+ domain, molecular replacement using models based on related proteins (such as PDB IDs 2R44, 3NBX and 4AKG and other, more remote homologs) as search models did not result in any solutions of sufficient quality for model building, possibly due to the C-terminal domain of CbbQ that is not found in any available search model. Using a mercury derivative we obtained phases which enabled us to build an atomic model into calculated density of the isomorphous native dataset. We refined the structure to Rwork/Rfree of 22.1/28.0% and 95% of the residues in the favored region of the Ramachandran plot (Table 2).
There are two CsoCbbQ molecules per asymmetric unit; expansion of crystal symmetry generates the full hexamer (Fig. 2b), consistent with the biologically active form of most AAA+ proteins and the size exclusion data (Supplementary Fig. 2). We were able to trace the protein backbone starting with residue R9 and, for one of the chains, extending to the C-terminus, with two loops missing between residues 79–89 and 153–159. One molecule of ADP is bound to each monomer, indicating that the ATP is hydrolyzed during crystallization. The two chains in the asymmetric unit align only moderately well (1.2 Å rmsd over all Cα atoms) with large differences observed in the C-terminal α-helices 8 and 9 (shifted by 3–4 Å) and with electron density for some loop regions missing in chain A. The differences observed could be explained by the inherent flexibility of the chains needed for the conformational changes coupled with ATP hydrolysis, which are typically transmitted to other proteins from AAA+ domain proteins.
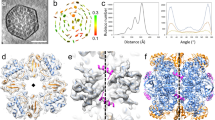
Structural overview of CsoCbbQ.
(a) Primary and secondary structure of H. nea CsoCbbQ. Labeling of the helices and sheets according to standard nomenclature for AAA+ proteins. Red tubes indicate α-helices, yellow arrows β-strands and dashed lines residues disordered in the CsoCbbQ structure. The C-terminal domain is indicated by grey letters. (b) Surface representation of the CsoCbbQ hexamer with alternating monomers shown in gold and grey and N- and C-terminal domains colored in darker and lighter tone, respectively. Inset shows the reverse side. The red asterisk indicates the location of the nucleotide binding site, which is not accessible to the surface (c) Cartoon representation of CsoCbbQ monomer rainbow colored from blue (N-) to red (C-) terminus; ADP is shown as red sticks. (d) Close-up of the nucleotide binding site with relevant residues labeled and shown as sticks. Arginine finger residue R168* is contributed by the neighboring chain.
Structurally, CsoCbbQ consists of the N-terminal P-loop NTPase domain (residues 1-175) characteristic of all AAA+ family proteins, comprising a beta sheet between two sets of α-helices (Fig. 2a,c). The order of the beta strands in the sheet is 5-1-4-3-2, typical for an AAA+ ATPase; α-helix 2 is interrupted by a partially disordered loop of 14 residues (G77-G91; Fig. 2a,c). The cyclic hexamer of N-terminal P-loop domains forms the central pore; the majority of inter-protomer contacts within the hexamer are formed by the NTPase domain. The N-terminal domain contains the characteristic Walker A (GXXXGK[T/S]) and Walker B (hhhhDE, h for hydrophobic residue) motifs (Fig. 2a,d) and is most closely related to the MoxR/dynein-related subfamily of AAA+ proteins (pfam07728). Sequence conservation is high throughout the entire NTPase domain (Supplementary Fig. 3) with the exception of the first 30 residues where only a PYY motif (residues 14–16) stands out. These tyrosine residues seem to be important for connecting the two subdomains; Y15 is involved in a π–π stacking interaction with the also highly conserved H188.
While the AAA+ domain of CbbQ can be structurally superimposed on its counterpart domain in CbbX, the C-terminal, CbbQ specific domain is structurally unique (Supplementary Fig. 4). The C-terminal domain (residues 176–270), the CbbQ-specific portion of the primary structure, forms a five-helix bundle that extends from the NTPase domain. (Fig. 2a,c). The CbbQ domain contributes only one residue, the conserved arginine finger residue R168, to the active site. Within the hexameric assembly, the majority of the CbbQ-specific domain is found on one surface of the hexamer (Fig. 2b), as a result this face of the hexamer is concave. Overall, the structure resembles a hexagon 105 Å in diameter (Fig. 2b). The pronounced hexagonal profile of CsoCbbQ is distinct from that of other structurally characterized AAA+ family members, which typically display a star-shaped outline31. This hexagonal profile of CsoCbbQ is a result of the gaps in the hexamer formed by the NTPase domain being filled by the five-helix bundles of each protomer (Fig. 2b). The central pore of CsoCbbQ is similar in size (19 Å) and charge to those of other AAA+ domain family members (Fig. 3b). The overall shape and straight edges of CbbQ are reminiscent of the CsoS1 protein hexamers which form the facets of the carboxysome shell. However, due to the different edge length (55 Å whereas in CsoS1A, a hexameric shell protein, the edges are 36 Å), it is not possible to readily fit CsoCbbQ into current models for the icosahedral carboxysome shell32.
Surface representations (left convex, right concave, which includes the majority of the CbbQ specific domain) of (a) Sequence conservation mapped to the surface of CsoCbbQ (yellow low, white intermediate and red high conservation). (b) Electrostatics of the hexamer from both sides, coloring from −4 kT/e (red) to +4 kT/e (blue).
CsoCbbQ is a component of the H. nea carboxysome
The occurrence of cbbQ in many chemoautotrophic α-carboxysome loci (Table 1) suggested a link between the CsoCbbQ paralog and the carboxysome. To investigate a potential physical association, purified H. nea carboxysomes were probed for the presence of CsoCbbQ. Co-migration of CsoCbbQ and the major carboxysome shell protein CsoS1 was investigated by immunoblotting18. The migration patterns of carboxysome-associated components can be monitored when sucrose gradient fractions of purified carboxysomes are loaded sequentially onto an SDS-PAGE gel and immunoblotted. Non-associated proteins sediment more slowly than carboxysomes during centrifugation, as shown in the gel image (Fig. 4a). The strongest CsoCbbQ and CsoS1 immunoblot signals are found in the same sucrose gradient fraction (#12), indicating CbbQ is associated with the H. nea carboxysome (Fig. 4a).
CsoCbbQ is associated with the carboxysome
(a) Immunoblots reveal co-migration of CsoCbbQ with the major carboxysome shell protein CsoS1 during sucrose gradient centrifugation. CbbQ = purified, hexa-histidine-tagged recombinant CbbQ, WC = wild type carboxysomes, 1–15 = sucrose gradient fractions from top (#1) to bottom (#15) of the gradient; (b) Immunoblot of purified carboxysomes (Whole); following breakage by freeze-thawing, the presence of CbbQ in the shell enriched (Pellet) and absence from the released protein fraction (Supernatant) suggests that CsoCbbQ interacts strongly with the shell.
To further define the interaction between CsoCbbQ and the carboxysome, isolated carboxysomes were subjected to a freeze/thaw cycle known to disrupt the H. nea carboxysome while keeping the shells largely intact16. Following centrifugation, fractured carboxysomes are separated into a shell-enriched pellet and a protein-enriched supernatant that consists mainly of released internal RuBisCO molecules. As seen in Fig. 4b, the major shell protein CsoS1 is found in the shell-enriched pellet, while RuBisCO immunoblots reveal that most of the RuBisCO protein is released into the supernatant (Fig. 4b). The presence of an anti-CsoCbbQ signal exclusively in the shell-enriched pellet fraction indicates CsoCbbQ is associated with the carboxysome shell (Fig. 4b).
Functional Characterization of CsoCbbQ
We established that CsoCbbQ has the ATPase activity predicted by the primary structure and from our crystal structure by spectrophotometrically measuring the production of ADP using a coupled assay with pyruvate kinase and lactate dehydrogenase33. CsoCbbQ demonstrated an average specific ATPase activity of 0.03 μmol ATP/min/mg CsoCbbQ at four different concentrations of CsoCbbQ (Supplementary Fig. 5). To determine if CsoCbbQ could function as a RuBisCO activase, the assay confirming the function of the red-type RuBisCO activase CbbX was undertaken with recombinant CsoCbbQ. We were unable to restore activity to H. nea RuBisCO inactivated by RubP (data not shown)34.
CsoCbbQ forms a complex with CsoCbbO
Based on the observation that CsoCbbQ and CbbO co-occur in a majority of non-cyanobacterial α-carboxysome loci and that MoxR AAA+ domain proteins such as CbbQ typically interact with proteins that contain VWA domains (as in CbbO), we investigated the potential for interaction between CsoCbbQ and the cso-associated CbbO (CsoCbbO). CsoCbbO proved insoluble when expressed in E. coli under a variety of conditions. However, when non-His6-tagged CsoCbbQ (NT-CbbQ) and His6-tagged csoCbbO were co-expressed, we were able to purify a complex (Fig. 5a,b). This complex eluted as a single peak in size-exclusion chromatography at approximately 254 kDa (Fig. 5c), most likely corresponding to one CsoCbbQ hexamer and one CsoCbbO subunit. The complex shows ATPase activity comparable to that of recombinant CsoCbbQ.
CsoCbbO/Q co-expression
(a) Solubility analysis of the IPTG-induced co-expression of CsoCbbQ (bottom arrows) and N-terminal histidine-tagged CsoCbbO (top arrows) (b) hisCsoCbbO was soluble when co-expressed with CsoCbbQ and could be purified as a CsoCbbO/Q complex using the affinity tag on CbbO. The difference in molecular weight between csoCbbQ in the complex and individually expressed recombinant CsoCbbQ is due to the absence of the His6-tag on CsoCbbQ in the complex. (c) SEC revealed the complex elutes as a single peak at approximately 254 kDa. Molecular weight standards used were thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa) and BSA (66 kDa).
Characterization of HncbbQ::Km mutant
To further attempt to identify the role of CsoCbbQ in carboxysome function, we generated a csocbbQ::Km knockout mutant. The endogenous carboxysomal csocbbQ gene was replaced by a kanamycin resistance cassette using homologous recombination as previously described35. This insertion was confirmed by PCR and genomic DNA sequencing. Mutant cells did not produce CsoCbbQ, as confirmed by immunoblotting of purified mutant carboxysomes (Fig. 6a).
Carboxysomes were purified from the csocbbQ::Km strain and examined by TEM (Fig. 6b). Structurally important carboxysome components can produce irregularly shaped carboxysomes when deleted36,37. However, wild type (WT) and mutant carboxysomes were of similar shape and diameter, indicating that, as previously observed for other low-abundance components15,16,36, csoCbbQ is not a crucial structural component. There was also no detectable phenotype as monitored by measuring the OD600 of batch cultures of WT and csocbbQ::Km grown in air and in 5% CO2, respectively, over a period of 40 hours (Supplementary Fig. 6). Mutant cells displayed no observable growth limitations, compared to the WT, when grown in air. Elimination of CsoCbbQ did not affect the size, number or spatial organization of carboxysomes; the ultrastructure of mutant cells was comparable to wildtype under both low and high CO2 growth conditions as judged by transmission electron microscopy (data not shown).
Discussion
We have determined the structure of CsoCbbQ, a AAA+ ATPase protein in its hexameric, nucleotide-bound form and show that it is an active ATPase. The occurrence of cbbQ near genes for Form I and Form II RuBisCO and prior evidence of its potential to affect the activity of recombinant RuBisCO in E. coli27,38 initially suggested two potential ATPase-associated functions for CsoCbbQ in the carboxysome: as an RbcX-like chaperone activity associated with RubisCO assembly and packaging or as an Rca (RuBisCO activase)/CbbX-like RuBisCO activase. In β-cyanobacteria, RuBisCO large and small subunit genes are frequently localized in an rbcLXS cluster with the Form I RuBisCO chaperone RbcX. This protein assembles as a homodimer that binds and stabilizes RbcL subunits during formation of the L8 core39. Inactivation of the rbcX gene in Synechococcus sp. 7002 resulted in a significant reduction in the amount of RbcL and RbcS subunits produced in vivo, while co-expression of RuBisCO subunits with RbcX in E. coli increased soluble hexadecamer formation and activity40. Such an effect on carboxysomal RuBisCO assembly in H. nea would be expected to yield reduced RuBisCO content in isolated mutant carboxysomes and likely a high CO2-requiring (hcr) phenotype in air-grown mutant cells, but no change in carboxysome polypeptide composition and morphology or growth rate and subcellular ultrastructure was observed in the csocbbQ mutant. However, the lack of a phenotype under “normal” growth conditions does not preclude a role for CsoCbbQ in RuBisCO assembly under specific (stress) conditions in vivo. No change in growth or RuBisCO content was observed in Synechococcus sp. PCC7942 rbcX::KmR mutants, despite the fact that the RbcX gene product was shown to positively affect RuBisCO assembly in E. coli41.
The second prospective ATPase related function for CsoCbbQ, as a RuBisCO activase, likewise could not be confirmed. There is a complete lack of structural homology between the C-terminal domains of CbbX and CsoCbbQ, suggesting the two proteins have distinct functions (Supplementary Fig. 4). CsoCbbQ does have relatively modest ATPase activity - approximately 30-50x lower than the reported in vitro rate for Rca from Arabidopsis (1.0–1.5 μmol ATP/min/mg), 20x lower than the red-type RuBisCO activase CbbX from Rhodobacter sphaeroides33,34 and 40× lower than the CbbQ from Hydrogenophilus thermoluteolus25. CsoCbbQ ATPase activity might be higher with its cognate substrate, which is not known at present. We were unable to determine a potential in vivo rate of ATP hydrolysis due to the low copy number of CsoCbbQ in isolated carboxysomes, but the observation of in vitro ATPase activity suggests the α-carboxysome may support an additional enzymatic reaction that could affect RuBisCO activation.
Attempts to determine the function of CsoCbbQ are complicated by the observation that it complexes with CsoCbbO. The function of CsoCbbO is even more enigmatic than that of CsoCbbQ. It is a large (788 amino acids in H. nea) protein in which the only discernible feature within the primary structure is a VWA domain in its C-terminus, a domain frequently found in proteins that are associated with AAA+ proteins. When co-expressed in E. coli, the two proteins form a complex and CsoCbbQ renders CsoCbbO soluble. The role of an ATP-hydrolyzing molecular chaperone for CsoCbbO would appear to suit CsoCbbQ, but further characterization is required.
Deletion of csocbbQ did not impair growth or carboxysome morphology in H. nea. However, there is a second cbbQ gene associated with that encoding Form II RuBisCO in H. nea. The Form II RuBisCO of Hydrogenovibrio marinus is expressed at elevated CO2 concentrations and can compensate for a reduction in Form IA RuBisCO activity under those conditions42. The form II-associated CbbQ would also likely be expressed under such conditions; it is unknown if this CbbQ gene product is Form II-specific or if it could compensate for the loss of CsoCbbQ activity under any conditions.
The lack of a discernible effect on carboxysome ultrastructure in the csocbbQ::Km mutant may be related to its low abundance; deletion of other minor components from the alpha carboxysome likewise does not have a discernible effect on carboxysome morphology15,16,36. The copy number of CsoCbbQ in the shell may indeed be flexible and, moreover, likely requires identifying a triggering stress for its functional importance to manifest. Elevated CO2 was the only environmental condition altered during growth experiments, but it is possible that further physiological stresses are required for CsoCbbQ to exert an effect on RuBisCO in the carboxysome. The position of csocbbQ (as well as csocbbO) in the expanded locus, downstream of the canonical cso operon, suggests that it is likely independently regulated, perhaps as a response to stress or the availability of nitrogen.
CbbQ is a member of the MoxR family of ATPases, many of which function as chaperones29,30. Some MoxR ATPases are known to interact with a protein containing a VWA domain (pfam00092), as found in the C-terminal region of CsoCbbO. Recently, subunits of the NADH:Ubiquinone Oxidoreductase (Nuo) I complex were shown to be additional interaction partners for a MoxR ATPase and its cognate VWA protein43. NuoL homologs (pfam00662 and pfam00361) are conserved in the typical expanded α-carboxysome loci (Fig. 1b,28); they encode membrane-spanning domains that have been implicated in inorganic carbon uptake44. In H. nea the gene encoding NuoL is followed by a putative transmembrane hypothetical protein, a 1046 amino acid protein DUF2309 (pfam10070), a Nitrogen Regulatory protein PII/SbtB homolog and a small hypothetical protein. The position of these genes between the verified interaction partners CsoCbbO and CsoCbbQ may indicate that their products play functional roles related to that of CsoCbbO and CsoCbbQ. In support of a functional relation among these gene products, the pfam00361 domain of NuoL is found in many proteins fused to pfam10070. Although speculative, in addition to co-expression, there may be physical interactions among these gene products of the expanded carboxysome locus that are important under some as yet to be identified conditions. This would account for the presumably different regulation of these genes relative to the canonical cso operon.
We have established that CsoCbbQ functions as an ATPase, as demonstrated both functionally (Supplementary Fig. 5) and structurally (Fig. 2d). Notably, the C-terminal domain (pfam08406) that is specific to CbbQ orthologs is a prominent feature of just one face of the hexamer (Fig. 2b). It consists of a bundle of five antiparallel helices, in contrast to most other AAA+ domain proteins that are composed of C-terminal four helix bundles. Structural homology searches using only the C-terminal domain result in only marginally scoring hits, with the best one being a ssRNA binding protein (PDB ID 2XC7) that has a sequence identity of 18% and an rmsd of 2.3 Å. This homology seems to be strictly structural though, since most of the residues binding the RNA are absent in CsoCbbQ. The highest conservation observed in the C-terminal domain is found where it is involved in intra- or inter-subunit contacts but also on the concave (CbbQ specific domain face) of the hexamer (Fig. 3a). Many of those residues are negatively charged (Fig. 3b). A possible explanation for this high conservation on the surface is that this region is important for interaction with a substrate or other protein(s), such as those forming the carboxysome shell or being part of a transmembrane carbon uptake complex.
The co-occurrence of CbbQ with RuBisCO in general and with the carboxysome in particular suggests a role possibly in either activation of RuBisCO or facilitating the packaging of RuBisCO into the α-carboxysome under some conditions. This is the first evidence for the association of an ATP-hydrolyzing enzyme with the carboxysome, as was speculated for the β-carboxysomes when Rca homologs that contained a domain putatively important for carboxysome localization were identified45. Additional biochemical and expression studies will be required to elucidate the precise function of CsoCbbQ; our negative results for activase activity may reflect the limitations of in vitro analysis outside of the confines of the carboxysome. The ATPase activity that we have identified and the novel AAA+ structure of CsoCbbQ will be valuable in interpretation of the results of further studies on CbbQ homologs.
Our results implicate CsoCbbQ as a component of the assembled carboxysome (Fig. 4a), where it is tightly associated with the shell (Fig. 4b). The striking hexagonal shape of the CsoCbbQ structure immediately evokes comparisons to carboxysome shell proteins, however, the CsoCbbQ hexamer is too large to be incorporated into existing models of carboxysome shell facets. The current working model of the carboxysome shell is based solely on structures of isolated shell protein hexamers (BMC-H) forming the facets and vertices formed by pentameric BMC-P32 with the geometric constraint of icosahedral symmetry. Clearly, this model is too simplistic; it is becoming increasingly apparent that there are additional protein subunits integrated into or tightly associated with the carboxysome shell. The abundant CsoS2 protein is a crucial component of the α-carboxysome shell13,46 and CsoSCA is so tightly associated with the shell that it cannot be purified without disintegrating the shell16,47. While CsoCbbQ may be too large to be integrated directly into the CsoS1 hexamer shell layer, the matching symmetry could enable it to dock onto a hexagonal shell protein. The association of the ATPase CsoCbbQ adds additional complexity to our current model of the carboxysome shell and suggests that it functions as more than an inert semi-permeable barrier.
Methods
Cloning and Expression
The csocbbQ (Hneap_0905) gene was amplified via PCR from H. neapolitanus genomic DNA with forward primer 5′-GGATCCATGACACAAAATGCAGATCAATATCG-3′ and reverse primer 5′-AAGCTTTTAAAAGAACGTTTTGACGACGG-3′ (italics represent added restriction enzyme sites). The amplified gene product was cloned into the pCR-BluntII-TOPO vector (Invitrogen) and sequenced. Positive clones were digested with restriction enzymes BamHI and HindIII (New England Biolabs) and ligated into linearized pCDFDuet-1 vector to give a pCDFDuet-csoCbbQ expression construct. IPTG-induced protein expression in chemically competent E. coli BL21(DE3) cells (Invitrogen) was carried out at 37 °C for 3 h, after which cells were lysed via three passes through a French Pressure cell. The crude extract was centrifuged and soluble CsoCbbQ in the supernatant was purified via His6-tag on Ni2+/NTA affinity resin (Thermo Scientific). Purified CsoCbbQ was eluted with 250 mM imidazole and was dialyzed twice in 4 l of 10 mM Tris-HCl, pH 8.0. Final protein concentrations were determined by BCA assay (Thermo Scientific/Pierce).
Shell association and ATPase activity
Co-migration and shell-association analysis was performed as described previously with a polyclonal primary antibody, generated by Cocalico Biologicals Inc., raised using affinity-purified his-CsoCbbQ for polyclonal antibody generation in rabbits, using the company’s standard inoculation protocol18. The rate of ATP hydrolysis was determined by spectrophotometrically measuring the rate of ADP formation in a coupled assay with pyruvate kinase and lactate dehydrogenase33. The reaction mixture contained 50 mM Tris-HCl pH 8.0, 20 mM KCl, 5 mM MgCl2, 2.5 mM ATP, 1 mM phosphoenolpyruvate, 0.3 mM NADH, 12 U/mL pyruvate kinase and 12 U/mL lactate dehydrogenase. Recombinant CbbQ protein was added at final concentrations of 0, 3.7, 7.4, 14.8 and 29.8 μM in a total reaction volume of 200 μL in a 96-well plate. The absorbance at 340 nm was measured in a Synergy 2 Multi-Mode plate reader (Bio-Tek) at 45 second time points for 15 minutes.
Mutant generation and characterization
The well-established protocol for generating carboxysome deletion mutants in H. nea was followed to yield csocbbQ::Km mutant cells35. A kanamycin resistance cassette was amplified with forward primer 5′-ttaaatcagaaagacggctacatcgaccagaacaaggcaatttaaCCGGAATTGCCAGCTGGG-3′ and reverse primer 5′-ctggtgtgtttcatgttgtgcgtattacctgttggtacggaaggcTCAGAAGAACTCGTCAAGAAGGCGATAG-3′. The lower case sequences represent regions of homology immediately upstream (forward primer) and downstream (reverse primer) of the endogenous csocbbQ gene and upper case sequences anneal to the Kmr cassette. Concurrently, a H. nea DNA fragment that encompasses the csocbbQ gene with adjacent 500 base pairs upstream and downstream was amplified from genomic DNA with forward primer 5′-GGATCCATCCGGAAACGGGCGATCG-3′ and reverse primer 5′-AAGCTTGCGATGATGTCGCAAGATAGG-3′. The resulting PCR product was cloned into the pCR-BluntII-TOPO vector (Invitrogen) and the sequence was verified. The csoCbbQ-500 region was then ligated into the BamHI and HindIII restriction sites of the pUC18 vector to yield pUC18-csoCbbQ500. Homologous recombination in E. coli DY330 cells was used to replace the csocbbQ gene in pUC18-csoCbbQ500 with the amplified Kmr cassette, utilizing the 45 bp of homology added during PCR. The resulting pUC18-CbbQ500-Kmr replacement construct was electroporated into exponentially growing H. nea cells as described previously for replacement of the endogenous csocbbQ gene with the kanamycin resistance cassette by homologous recombination in vivo35.
Successful replacement of the csocbbQ gene with the Kmr resistance cassette was confirmed by genomic DNA sequencing. A culture of mutant cells was maintained in a Multifors 1-L chemostat (Infors HT) with kanamycin-supplemented medium at 30°C. Carboxysomes were isolated from chemostat-grown cells as described previously and the absence of CsoCbbQ in mutant carboxysomes was confirmed by immunoblotting48. Ultrastructure of isolated mutant carboxysomes was examined by transmission electron microscopy (TEM). A 1.0 mg/mL carboxysome sample was loaded onto a 300-mesh Formvar-coated copper grid and stained with 1% ammonium molybdate, pH 8.0. Images were acquired with a Zeiss 900 TEM with a Model 785 ES1000W Erlangshen CCD camera at 140,000× magnification. Growth of mutant cells in air and 5% CO2 at 30 °C was monitored by measuring the OD600 of 100 ml batch cultures grown in Kanamycin-free media at several time points over a period of 42 hours. Growth of WT cells under identical conditions was monitored as a control.
Complex characterization
The Duet vector expression system (Novagen) was used to co-express His6-tagged CsoCbbO (HT-CbbO) and CsoCbbQ without an affinity tag (NT-CbbQ). A pCDFDuet-NTCbbQ expression construct was generated by PCR amplification with the forward primer 5′-CCATGGATGACACAAAATGCAGATCAATATCG-3′, which allowed insertion of the gene sequence into the pCDFDuet-1 NcoI restriction site located upstream of the hexa-histidine sequence in the vector and the reverse primer listed previously. The csocbbO (Hneap_0910) gene was amplified via PCR from H. nea genomic DNA with forward primer 5′-GGATCCGATGAACCCAGCGACTGAA-3′ and reverse primer 5′-AAGCTTCTATCGCGTCATCGACAAAT-3′. The amplified gene product was cloned as described previously and was ligated into the BamHI and HindIII sites of the pETDuet-1 vector. Plasmids pETDuet-HTCbbO and pCDFDuet-NTCbbQ were co-transformed into E. coli BL21(DE3) chemically competent cells and the transformants grown on LB-agar medium supplemented with ampicillin and spectinomycin. Expression and purification of the complex followed the same procedure described for recombinant CsoCbbQ. The molecular weight of the complex was estimated by size exclusion chromatography (SEC) on a prepacked Superdex 200 10/300 GL column (GE Life Sciences). A protein standard solution of thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa) and BSA (66 kDa) (Sigma-Aldrich) was used for molecular weight determination; a blue dextran solution was used to determine the column void volume. Runs were performed with the BioLogic DuoFlow System (Bio-Rad). Sample injection volumes were 100 μl and a de-gassed buffer solution of 10 mM Tris-HCl, pH 8.0, with 100 mM NaCl was used as the column eluent at a flow rate of 0.25 ml/min. Absorbance was measured by a BioLogic QuadTec UV-Vis Detector (Bio-Rad). The CsoCbbO:CsoCbbQ ratio of the purified complex was determined by spot densitometry analysis with Quantity One® software in a Bio-Rad 4000 MP Imaging system, following SDS-PAGE separation of peak fractions collected during SEC.
Bioinformatics
Chemoautotrophs with cso operon gene clusters were selected by combining search results from non-redundant homology searches with the Basic Local Alignment Search Tool (BLAST) algorithm and top homolog hits on the Joint Genome Institute Integrated Microbial Genome (JGI-IMG) database, using the α-carboxysome-specific CsoS2 peptide as a diagnostic marker. A Bit Score of 100 was used as a cut-off value. After discarding the α-cyanobacteria, 32 chemoautotrophs were examined for co-occurrence of cbbO and cbbQ genes using the gene ortholog neighborhood function in the JGI-IMG database. The sequence logo was generated with Weblogo49 and the phylogenetic tree was made with PhyML50.
Crystallization and structure determination
CsoCbbQ was crystallized at 22°C at concentrations of 5–10 mg/ml in 0.1 M HEPES pH 7.5, 6% PEG-8000, 8% ethylene glycol in sitting drop trays. Crystals were stabilized by adding 15% ethylene glycol to the crystallization drop before flash freezing in liquid nitrogen. Heavy atom derivatives were obtained by adding 0.1 μl of a 10 mM solution of Thiomersal (Ethyl (2-mercaptobenzoato-(2-)-O,S) mercurate(1-) sodium) to a 10 μl drop containing the stabilized crystals. Diffraction data were collected at beam line 5.0.2 of the Advanced Light Source of the Lawrence Berkeley National Lab. Diffraction data were integrated with XDS51 and scaled with SCALA (CCP4)52. The structure of CsoCbbQ was solved by locating the mercury modified cysteines using phenix.phaser53 and using those sites to generate an initial electron density map. Automatic building of a model into the density using buccaneer52 was followed by manual rebuilding/refinement cycles using COOT54 and phenix.refine. Statistics for diffraction data collection, structure determination and refinement are summarized in Table 2. Figures of crystal structures were prepared using pymol (www.pymol.org) molecular visualization software.
Additional Information
How to cite this article: Sutter, M. et al. Structural Characterization of a Newly Identified Component of α-Carboxysomes: The AAA+ Domain Protein CsoCbbQ. Sci. Rep. 5, 16243; doi: 10.1038/srep16243 (2015).
References
Ellis, R. J. Biochemistry: Tackling unintelligent design. Nature 463, 164–165, doi: 10.1038/463164a (2010).
Badger, M. R. & Bek, E. J. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. Journal of experimental botany 59, 1525–1541, doi: 10.1093/jxb/erm297 (2008).
Parry, M. A., Andralojc, P. J., Mitchell, R. A., Madgwick, P. J. & Keys, A. J. Manipulation of Rubisco: the amount, activity, function and regulation. Journal of experimental botany 54, 1321–1333 (2003).
Price, G. D., Badger, M. R., Woodger, F. J. & Long, B. M. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. Journal of experimental botany 59, 1441–1461, doi: 10.1093/jxb/erm112 (2008).
Shively, J. M., English, R. S., Baker, S. H. & Cannon, G. C. Carbon cycling: the prokaryotic contribution. Current opinion in microbiology 4, 301–306 (2001).
Dobrinski, K. P., Longo, D. L. & Scott, K. M. The carbon-concentrating mechanism of the hydrothermal vent chemolithoautotroph Thiomicrospira crunogena. Journal of bacteriology 187, 5761–5766, doi: 10.1128/JB.187.16.5761-5766.2005 (2005).
Hopkinson, B. M., Young, J. N., Tansik, A. L. & Binder, B. J. The minimal CO2-concentrating mechanism of Prochlorococcus spp. MED4 is effective and efficient. Plant physiology 166, 2205–2217, doi: 10.1104/pp.114.247049 (2014).
Yeates, T. O., Kerfeld, C. A., Heinhorst, S., Cannon, G. C. & Shively, J. M. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nature reviews. Microbiology 6, 681–691, doi: 10.1038/nrmicro1913 (2008).
Dou, Z. et al. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. The Journal of biological chemistry 283, 10377–10384, doi: 10.1074/jbc.M709285200 (2008).
Rae, B. D., Long, B. M., Badger, M. R. & Price, G. D. Functions, compositions and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiology and molecular biology reviews: MMBR 77, 357–379, doi: 10.1128/MMBR.00061-12 (2013).
Cannon, G. C. et al. Organization of carboxysome genes in the thiobacilli. Current microbiology 46, 115–119, doi: 10.1007/s00284-002-3825-3 (2003).
Cai, F., Heinhorst, S., Shively, J. M. & Cannon, G. C. Transcript analysis of the Halothiobacillus neapolitanus cso operon. Archives of microbiology 189, 141–150, doi: 10.1007/s00203-007-0305-y (2008).
Cai, F. et al. Advances in Understanding Carboxysome Assembly in Prochlorococcus and Synechococcus Implicate CsoS2 as a Critical Component. Life 5, 1141–1171, doi: 10.3390/life5021141 (2015).
Sawaya, M. R. et al. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. The Journal of biological chemistry 281, 7546–7555, doi: 10.1074/jbc.M510464200 (2006).
So, A. K. et al. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. Journal of bacteriology 186, 623–630 (2004).
Heinhorst, S. et al. Characterization of the carboxysomal carbonic anhydrase CsoSCA from Halothiobacillus neapolitanus. Journal of bacteriology 188, 8087–8094, doi: 10.1128/JB.00990-06 (2006).
Klein, M. G. et al. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. Journal of molecular biology 392, 319–333, doi: 10.1016/j.jmb.2009.03.056 (2009).
Roberts, E. W., Cai, F., Kerfeld, C. A., Cannon, G. C. & Heinhorst, S. Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. Journal of bacteriology 194, 787–795, doi: 10.1128/JB.06444-11 (2012).
Eisenhut, M. et al. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant physiology 144, 1946–1959, doi: 10.1104/pp.107.103341 (2007).
Wheatley, N. M., Sundberg, C. D., Gidaniyan, S. D., Cascio, D. & Yeates, T. O. Structure and identification of a pterin dehydratase-like protein as a ribulose-bisphosphate carboxylase/oxygenase (RuBisCO) assembly factor in the alpha-carboxysome. The Journal of biological chemistry 289, 7973–7981, doi: 10.1074/jbc.M113.531236 (2014).
Kolesinski, P., Belusiak, I., Czarnocki-Cieciura, M. & Szczepaniak, A. Rubisco Accumulation Factor 1 from Thermosynechococcus elongatus participates in the final stages of ribulose-1,5-bisphosphate carboxylase/oxygenase assembly in Escherichia coli cells and in vitro. The FEBS journal 281, 3920–3932, doi: 10.1111/febs.12928 (2014).
Whitney, S. M., Birch, R., Kelso, C., Beck, J. L. & Kapralov, M. V. Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proceedings of the National Academy of Sciences of the United States of America 112, 3564-3569, doi: 10.1073/pnas.1420536112 (2015).
Snider, J. & Houry, W. A. AAA+ proteins: diversity in function, similarity in structure. Biochemical Society transactions 36, 72–77, doi: 10.1042/BST0360072 (2008).
Lupas, A. N. & Martin, J. AAA proteins. Current opinion in structural biology 12, 746–753 (2002).
Hayashi, N. R. & Igarashi, Y. ATP binding and hydrolysis and autophosphorylation of CbbQ encoded by the gene located downstream of RubisCO genes. Biochemical and biophysical research communications 290, 1434–1440, doi: 10.1006/bbrc.2002.6366 (2002).
Hayashi, N. R., Arai, H., Kodama, T. & Igarashi, Y. The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochemical and biophysical research communications 241, 565–569, doi: 10.1006/bbrc.1997.7853 (1997).
Hayashi, N. R., Arai, H., Kodama, T. & Igarashi, Y. The cbbQ genes, located downstream of the form I and form II RubisCO genes, affect the activity of both RubisCOs. Biochemical and biophysical research communications 265, 177–183, doi: 10.1006/bbrc.1999.1103 (1999).
Axen, S. D., Erbilgin, O. & Kerfeld, C. A. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS computational biology 10, e1003898, doi: 10.1371/journal.pcbi.1003898 (2014).
Ammelburg, M., Frickey, T. & Lupas, A. N. Classification of AAA+ proteins. Journal of structural biology 156, 2–11, doi: 10.1016/j.jsb.2006.05.002 (2006).
Snider, J. & Houry, W. A. MoxR AAA+ ATPases: a novel family of molecular chaperones? Journal of structural biology 156, 200–209, doi: 10.1016/j.jsb.2006.02.009 (2006).
De Mot, R., Nagy, I., Walz, J. & Baumeister, W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends in microbiology 7, 88–92 (1999).
Tanaka, S. et al. Atomic-level models of the bacterial carboxysome shell. Science. 319, 1083–1086, doi: 10.1126/science.1151458 (2008).
Robinson, S. P. & Portis, A. R., Jr. Adenosine triphosphate hydrolysis by purified rubisco activase. Archives of biochemistry and biophysics 268, 93–99 (1989).
Mueller-Cajar, O. et al. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199, doi: 10.1038/nature10568 (2011).
English, R. S., Jin, S. & Shively, J. M. Use of Electroporation To Generate a Thiobacillus neapolitanus Carboxysome Mutant. Applied and environmental microbiology 61, 3256–3260 (1995).
Cai, F. et al. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PloS one 4, e7521, doi: 10.1371/journal.pone.0007521 (2009).
Price, G. D. & Badger, M. R. Isolation and Characterization of High CO(2)-Requiring-Mutants of the Cyanobacterium Synechococcus PCC7942: Two Phenotypes that Accumulate Inorganic Carbon but Are Apparently Unable to Generate CO(2) within the Carboxysome. Plant physiology 91, 514–525 (1989).
Parry, M. A., Keys, A. J., Madgwick, P. J., Carmo-Silva, A. E. & Andralojc, P. J. Rubisco regulation: a role for inhibitors. Journal of experimental botany 59, 1569–1580, doi: 10.1093/jxb/ern084 (2008).
Saschenbrecker, S. et al. Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 129, 1189–1200, doi: 10.1016/j.cell.2007.04.025 (2007).
Onizuka, T. et al. The rbcX gene product promotes the production and assembly of ribulose-1,5-bisphosphate carboxylase/oxygenase of Synechococcus sp. PCC7002 in Escherichia coli. Plant & cell physiology 45, 1390–1395, doi: 10.1093/pcp/pch160 (2004).
Baker, S. H., Jin, S., Aldrich, H. C., Howard, G. T. & Shively, J. M. Insertion mutation of the form I cbbL gene encoding ribulose bisphosphate carboxylase/oxygenase (RuBisCO) in Thiobacillus neapolitanus results in expression of form II RuBisCO, loss of carboxysomes and an increased CO2 requirement for growth. Journal of bacteriology 180, 4133–4139 (1998).
Yoshizawa, Y., Toyoda, K., Arai, H., Ishii, M. & Igarashi, Y. CO2-responsive expression and gene organization of three ribulose-1,5-bisphosphate carboxylase/oxygenase enzymes and carboxysomes in Hydrogenovibrio marinus strain MH-110. Journal of bacteriology 186, 5685–5691, doi: 10.1128/JB.186.17.5685-5691.2004 (2004).
Wong, K. S. et al. The MoxR ATPase RavA and its cofactor ViaA interact with the NADH:ubiquinone oxidoreductase I in Escherichia coli. PloS one 9, e85529, doi: 10.1371/journal.pone.0085529 (2014).
Gaudana, S. B., Zarzycki, J., Moparthi, V. K. & Kerfeld, C. A. Bioinformatic analysis of the distribution of inorganic carbon transporters and prospective targets for bioengineering to increase C uptake by cyanobacteria. Photosynthesis research, doi: 10.1007/s11120-014-0059-8 (2014).
Zarzycki, J., Axen, S. D., Kinney, J. N. & Kerfeld, C. A. Cyanobacterial-based approaches to improving photosynthesis in plants. Journal of experimental botany 64, 787–798, doi: 10.1093/jxb/ers294 (2013).
Baker, S. H. et al. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in thiobacillus neapolitanus. Archives of microbiology 172, 233–239 (1999).
Heinhorst, S., Cannon, G. & Shively, J. In Complex Intracellular Structures in Prokaryotes Vol. 2 Microbiology Monographs (ed JessupM Shively) Ch. 23, 141–165, (Springer Berlin Heidelberg, 2006).
Cannon, G. C. & Shively, J. M. Characterization of a homogeneous preparation of carboxysomes from Thiobacillus neapolitanus. Archives of microbiology 134, 52–59 (1983).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome research 14, 1188–1190, doi: 10.1101/gr.849004 (2004).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology 59, 307–321, doi: 10.1093/sysbio/syq010 (2010).
Kabsch, W. XDS Acta crystallographica. Section D, Biological crystallography 66, 125–132, doi: 10.1107/S0907444909047337 (2010).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67, 235–242, doi: 10.1107/S0907444910045749 (2011).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D 68, 352–367, doi: Doi 10.1107/S0907444912001308 (2012).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography 60, 2126–2132, doi: 10.1107/S0907444904019158 (2004).
Acknowledgements
Support for this work was provided by the US Department of Energy, Basic Energy Sciences, DE-FG02-91ER20021 with infrastructure support from MSU AgBio Research (CAK, CRG and MS) and by National Science Foundation grant MCB-1244534 (SH and GCC). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
G.C.C., S.H. and C.A.K. designed the study. M.S., E.W.R., S.H. and C.A.K. analyzed the data and wrote the manuscript. E.W.R. performed the cloning, expression, mutant studies, protein purification and protein assays. M.S. crystallized the protein and solved the structure. R.C.G. performed T.E.M. analysis. C.B. prepared cells for T.E.M. analysis. S.D. created the CbbQ expression construct and performed protein interaction studies; K.L. created the deletion mutant construct. All authors analyzed the results and approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sutter, M., Roberts, E., Gonzalez, R. et al. Structural Characterization of a Newly Identified Component of α-Carboxysomes: The AAA+ Domain Protein CsoCbbQ. Sci Rep 5, 16243 (2015). https://doi.org/10.1038/srep16243
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep16243
This article is cited by
-
Bacterial microcompartments
Nature Reviews Microbiology (2018)
-
Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts
Nature Communications (2018)
-
Permanent draft genome of Thiobacillus thioparus DSM 505T, an obligately chemolithoautotrophic member of the Betaproteobacteria
Standards in Genomic Sciences (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.