Abstract
Studies of hair cell regeneration in the postnatal cochlea rely on fate mapping of supporting cells. Here we characterized a Sox2-CreER knock-in mouse line with two independent reporter mouse strains at neonatal and mature ages. Regardless of induction age, reporter expression was robust, with CreER activity being readily detectable in >85% of supporting cells within the organ of Corti. When induced at postnatal day (P) 28, Sox2-CreER activity was exclusive to supporting cells demonstrating its utility for fate mapping studies beyond this age. However, when induced at P1, Sox2-CreER activity was also detected in >50% of cochlear hair cells, suggesting that Sox2-CreER may not be useful to fate map a supporting cell origin of regenerated hair cells if induced at neonatal ages. Given that this model is currently in use by several investigators for fate mapping purposes and may be adopted by others in the future, our finding that current protocols are effective for restricting CreER activity to supporting cells at mature but not neonatal ages is both significant and timely.
Similar content being viewed by others
Introduction
Mammals have limited capacity to regenerate cochlear hair cells after ototoxic damage1,2,3,4,5,6,7, whereas non-mammalian vertebrates can spontaneously regenerate damaged hair cells from surrounding supporting cells by either direct transdifferentiation or mitotic regeneration8,9,10,11,12,13,14. Various genetic and therapeutic approaches have been taken to regenerate hair cells in the postnatal mammalian cochlea. However, in studies where experimental animals appear to possess greater numbers of hair cells than similarly damaged controls, it is often difficult to discriminate between the generation of new hair cells from underlying supporting cells versus the preservation of existing hair cells via intra-cellular repair and/or pro-survival mechanisms15,16,17,18,19,20,21,22. Indeed, in many studies where induced regeneration has been attempted in mammals, the numbers of hair cells are often less than that of an undamaged cochlea and any functional benefit often falls dramatically short of normal hearing1,17,18,23,24,25,26,27, suggesting that a mitigation of hair cell loss and functional recovery of surviving cells is as plausible an explanation as regeneration. Furthermore, several studies demonstrate that a single manipulation (e.g. the addition of a trophic factor or the expression of Atoh1) can act to promote either the phenotypic conversion of supporting cells to hair cell-like cells or the survival of residual hair cells, thus directly illustrating the difficulty in parsing these two putative mechanisms19,21,28,29,30,31. Therefore, it has become critical to demonstrate that newly regenerated hair cells are derived from supporting cells as in non-mammalian vertebrates. To accomplish this traditionally difficult task, investigators are now relying on genetic fate mapping methods where supporting cells specifically are transiently induced to express a permanent marker prior to hair cell damage, so that only subsequently regenerated hair cells derived from supporting cells would also express this marker. Such fate mapping is now commonly performed using CreER-mediated reporter expression in transgenic mice where transient tamoxifen administration allows CreER molecules to enter the nucleus and excise premature stop codons thus enabling permanent expression of a reporter. By utilizing a CreER transgene that possesses a supporting cell-specific promoter, or by knocking the CreER transgene into the locus of a gene that is normally only expressed in supporting cells, permanent reporter expression can be acutely induced and retained only in cells that were supporting cells at the time of tamoxifen administration. We and others have previously demonstrated several mouse lines that are useful for this purpose (Prox1-CreER, Fgfr3-iCreER, Plp-CreER and Glast-CreER)5,26,27,32,33; however, these reporters tend to be limited in that they do not allow for fate mapping of all of the supporting cells, but rather are only specific to subsets of supporting cell types.
The Sox2-CreER knock-in mouse line represents a promising model for the fate mapping of supporting cells since Sox2 is expressed in all of the supporting cells, not just certain supporting cell subtypes2,25,32,34,35,36. However, several lines of evidence suggest the presence of Sox2 expression / transcriptional activity in cochlear hair cells at postnatal and adult ages2,27,37,38,39,40,41. It is therefore critical to validate at what age, if any, that tamoxifen induction of this Sox2-CreER line will label only supporting cells and not hair cells. Finding an appropriate induction age is critical since the induction of reporter expression in any existing hair cells at the time of tamoxifen induction would defeat the purpose of such fate mapping and could not then be effectively used to support the conclusion that newly regenerated hair cells are derived from supporting cells in hair cell regeneration studies. Additionally, the vast majority of regeneration studies in the mouse cochlea are undertaken at neonatal ages due to greater plasticity of the cochlea and better survival of the tissue in culture at younger ages1,2,5,6,20,26,27,36,42,43,44,45,46,47,48,49,50,51.
Here we characterized the Sox2-CreER line at two ages, P1 and P28, by using two independent reporter mouse lines (Ai14 and mT/mG). Ai14 is a highly sensitive Cre reporter line where the tdTomato reporter gene, following Cre mediated excision of the loxP-Stop-loxP cassette, is driven by the Rosa26-CMV β-actin enhancer (CAG) promoter5,32,33,52,53. The mT/mG line is also driven by the Rosa26-CAG promoter, which is then followed by a loxP-mtdTomato-Stop-loxP–mGFP cassette. In the mT/mG mice, Cre negative cells, or uninduced Cre-positive cells, constitutively express mtdTomato, but upon Cre or CreER mediated excision of the mtdTomato, mGFP instead is expressed54. In this latter model, both mtdTomato and mGFP are membrane bound, while tdTomato in the Ai14 reporter line is mainly cytoplasmic and nuclear.
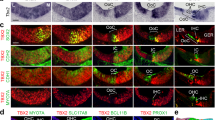
To test the supporting cell specificity of the Sox2-CreER line, we induced Sox2-CreER; Ai14 mice via two intraperitoneal tamoxifen injections (75 mg/kg body weight) spaced 24 hours apart between P0-P1 and analyzed the cochleae at P8 (Fig. 1A,B). Immunostaining of myosin-VIIa (Myo7a) and Sox2 allowed us to label hair cells and supporting cells, respectively, while the fluorescent reporter (tdTomato) was visualized directly using fluorescent, laser scanning confocal microscope. We quantified cell numbers of each supporting cell subtype and inner or outer hair cell subtype based on cell locations and immunolocalization of Myo7a and Sox2 in these architecturally organized organs of Corti. As expected, we found that a vast majority (>85%) of supporting cells are double positive for the Cre-activated reporter (tdTomato) and Sox2 (Fig. 1A,B); however, we also found that ~50% of Myo7a positive hair cells were also labeled by the Cre-activated tdTomato reporter (Fig. 1A,B). This was most noticeable in the apical turns of the cochleae, where 83.92 ± 10.32% of Myo7a positive hair cells were also tdTomato positive, as compared to 36.15 ± 19.41% in the middle turns and 19.78 ± 16.47% in the basal turns.
A,B. Sox2-CreER labels >85% supporting cells and >50% inner and outer hair cells in the organ of Corti when induced at P0-1. C,D. Sox2CreER labels only supporting cells in the organ of Corti when induced at P28. Wholemount cochlear confocal images (A,C) and quantification (B,D) of Sox2-CreER; ROSA-CAG-Stop-floxed-tdTomato (Ai14) when induced at P0-P1 and analyzed at P8, or induced at P28 and analyzed at P35. Hair cells are labeled with Myo7a antibody (white) in the P0-P1 induced samples (A) and in the P28 induced samples (C). Sox2 immunostaining (green) labels supporting cells and tdTomato (red) labels Cre activity. (B and D). Percentages of tdTomato+ cells in each subtype of supporting cells (Inner phallengeal cells [Iph], Inner pillar cells [IPC], outer pillar cells [OPC], Deiters’ cells [DC], Hensen cells [Hens]) and hair cells (HCs). Scale bars = 20 μm. Error bars = ±1 standard error in three independent mice.
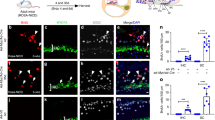
Similarly, we analyzed the cochleae of Sox2-CreER; mT/mG mice at P38 (Fig. 2) after tamoxifen (75 mg/kg body weight) had been administered by a single intraperitoneal injection at P125. In the apical turns, we found that only ~40% of the Myo7a positive hair cells remained mtdTomato positive, whereas nearly 60% of the Myo7a positive hair cells were mGFP positive, suggesting Sox2-CreER activity. These results are consistent with what we found in Sox2-CreER; Ai14 mice (Fig. 1A,B) and suggest that neonatal tamoxifen induction in the Sox2-CreER line may not be a suitable approach for fate mapping cochlear supporting cells.
Sox2-CreER labels both supporting cells and hair cells when induced at P1.
Tamoxifen was intraperitoneally injected into Sox2-CreER; mTmG mice at P1 and cochleae were analyzed at P38. A. The whole mount of apical cochlear turn with mGFP (green) and mtdTomato (red) fluorescence and Myo7a immunostaining (blue) using LSM700 confocal laser scanning microscope. B. The optical xz-plane of the line in A. Arrowheads label mGFP+ hair bundles. Scale = 20 μm. C. Percentages of mGFP+ hair cells in a 300-μm region from the apical turn. Error bars = ±1 standard error, N = 3 independent mice.
In contrast, we also injected Sox2-CreER; Ai14 mice with tamoxifen (250 mg/kg body weight) by a single injection at P28 and analyzed the cochleae at P35. Under these conditions, we found that many (>85%) Sox2 positive supporting cells, but no hair cells (immunostained for Myo7a), were labeled with the Cre-activated reporter (tdTomato) (Fig. 1C,D).
Our studies demonstrate that the Sox2-CreER line can be used as an effective genetic tool for the fate mapping of cochlear supporting cells following mature, but not neonatal tamoxifen induction. In support, Sox2 has been demonstrated to be expressed in progenitor cells, developing hair cells and supporting cells in late mouse embryos35,38,39,55,56,57 and while it is clearly down-regulated to some extent in differentiating hair cells between E16.5 and P4, several reports suggest that Sox2 is still expressed in cochlear hair cells at postnatal ages2,27,37,38,39,40,41. Our results here, using two independent fate mapping reporters, strongly support the hypothesis that Sox2 is indeed expressed in cochlear hair cells at P0-P1. This is of significance since several previously published studies have suggested the use of the Sox2-CreER line to fate map presumptive supporting cells following P1 induction2,25,36. While one of these reports has recently been corrected to clarify that the induction was carried out at P21 rather than P125, it is likely that investigators may still seek to use this mouse line to lineage trace supporting cells. The data presented here, which are corroborated by a previous report using the mT/mG reporter2, contradict that notion, suggesting limited or no utility for fate mapping since such a large number of cochlear hair cells exhibit Cre activity in response to neonatal induction of Sox2-CreER. However, this does not completely negate the usefulness of the Sox2-CreER mouse line for the fate mapping of supporting cells as the data also show that the Sox2-CreER line is specific to and robustly expressed in, cochlear supporting cells following induction at later (i.e. young adult) ages.
Methods
Animals
Mice were housed under a 12 h light/dark cycle with free access to food and water. The Animal Care and Use Committees of St. Jude Children’s Research Hospital approved all of the protocols performed in this study and the methods were carried out in accordance with the approved guidelines. Sox2-CreER mice were generously provided by Konrad Hochedlinger, Harvard University and are now available from Jackson Laboratory (Stock #017593). The mT/mG (Stock# 007576) mice were purchased from The Jackson Laboratory. Genotyping of Sox2-CreER and mT/mG mice were previously described34,54.
Histological analysis
In order to look at mGFP and mtdTomato distributions in organ of Corti, the Sox2-CreER; mT/mG mice were anesthetized by intraperitoneal injection with Avertin (0.5 mg/kg body weight) and fixed transcardially using fixative solution (4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4)). The inner ears were perfused via round window and fixed using the fixative solution at room temperature for 4 hours. After the fixation, the cochleae were carefully dissected out. Primary antibodies used were rabbit anti-myo7a (Proteus Bioscience, at 1:100), goat anti-Sox2 (Santa Cruz, at 1:500). The immunofluorescence was visualized by adding Alexa Fluor 405 goat anti-rabbit IgG (H + L), or Alexa Fluor 488 donkey anti-goat IgG (H + L) and Alexa Fluor 647 donkey anti-rabbit IgG (H + L) (Molecular Probes, Eugene, OR, USA). Fluorescence images were taken from N = 3 Sox2-CreER-Ai14 mice at P8 (following P0-P1 induction), N = 4 Sox2-CreER;Ai14 mice at P35 (following P28 induction) and N = 3 Sox2-CreER;mT/mG mice at P38 (following P1 induction). Images were acquired with a Zeiss Axiophot2 microscope equipped with a 40× oil immersion and 1.4 NA objective using a LSM700 confocal laser scanning image system (Carl Zeiss, Jena, Germany). For the Sox2-CreER;Ai14 mice, hair cells and supporting cells were counted from six representative images per cochlea, two from each turn (apical, middle and basal). For the Sox2-CreER;mT/mG mice, cells were counted from two representative images per cochlea, both in the apical turn.
Additional Information
How to cite this article: Walters, B. J. et al. Sox2-CreER mice are useful for fate mapping of mature, but not neonatal, cochlear supporting cells in hair cell regeneration studies. Sci. Rep. 5, 11621; doi: 10.1038/srep11621 (2015).
References
Cox, B. C. et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816–829 (2014).
Bramhall, N. F., Shi, F., Arnold, K., Hochedlinger, K. & Edge, A. S. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep 2, 311–322 (2014).
Oshima, K. et al. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol 8, 18–31 (2007).
Oesterle, E. C., Chien, W. M., Campbell, S., Nellimarla, P. & Fero, M. L. p27(Kip1) is required to maintain proliferative quiescence in the adult cochlea and pituitary. Cell Cycle 10, 1237–1248 (2011).
Mellado Lagarde, M. M. et al. Spontaneous regeneration of cochlear supporting cells after neonatal ablation ensures hearing in the adult mouse. Proc Natl Acad Sci USA 111, 16919–16924 (2014).
Chai, R. et al. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci USA 109, 8167–8172 (2012).
White, P. M., Doetzlhofer, A., Lee, Y. S., Groves, A. K. & Segil, N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984–987 (2006).
Lush, M. E. & Piotrowski, T. Sensory hair cell regeneration in the zebrafish lateral line. Dev Dyn 243, 1187–1202 (2014).
Corwin, J. T. & Cotanche, D. A. Regeneration of sensory hair cells after acoustic trauma. Science 240, 1772–1774 (1988).
Ryals, B. M. & Rubel, E. W. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science 240, 1774–1776 (1988).
Warchol, M. E. & Corwin, J. T. Regenerative proliferation in organ cultures of the avian cochlea: identification of the initial progenitors and determination of the latency of the proliferative response. J Neurosci 16, 5466–5477 (1996).
Baird, R. A., Steyger, P. S. & Schuff, N. R. Mitotic and nonmitotic hair cell regeneration in the bullfrog vestibular otolith organs. Ann N Y Acad Sci 781, 59–70 (1996).
Adler, H. J. & Raphael, Y. New hair cells arise from supporting cell conversion in the acoustically damaged chick inner ear. Neurosci Lett 205, 17–20 (1996).
Jones, J. E. & Corwin, J. T. Regeneration of sensory cells after laser ablation in the lateral line system: hair cell lineage and macrophage behavior revealed by time-lapse video microscopy. J Neurosci 16, 649–662 (1996).
Kopke, R. D. et al. Growth factor treatment enhances vestibular hair cell renewal and results in improved vestibular function. Proc Natl Acad Sci USA 98, 5886–5891 (2001).
Gale, J. E., Meyers, J. R., Periasamy, A. & Corwin, J. T. Survival of bundleless hair cells and subsequent bundle replacement in the bullfrog’s saccule. J Neurobiol 50, 81–92 (2002).
Izumikawa, M. et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med 11, 271–276 (2005).
Hori, R. et al. Pharmacological inhibition of Notch signaling in the mature guinea pig cochlea. Neuro report 18, 1911–1914 (2007).
Hayashi, Y., Yamamoto, N., Nakagawa, T. & Ito, J. Insulin-like growth factor 1 inhibits hair cell apoptosis and promotes the cell cycle of supporting cells by activating different downstream cascades after pharmacological hair cell injury in neonatal mice. Mol Cell Neurosci 56, 29–38 (2013).
Li, W. et al. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc Natl Acad Sci USA 112, 166–171 (2015).
Feghali, J. G. et al. Mammalian auditory hair cell regeneration/repair and protection: a review and future directions. Ear, Nose, & Throat J 77, 276, 280, 282-275 (1998).
Du, X. et al. Regeneration of mammalian cochlear and vestibular hair cells through Hes1/Hes5 modulation with siRNA. Hear Res 304, 91–110 (2013).
Kraft, S., Hsu, C., Brough, D. E. & Staecker, H. Atoh1 induces auditory hair cell recovery in mice after ototoxic injury. Laryngoscope 123, 992–999 (2013).
Atkinson, P. J., Wise, A. K., Flynn, B. O., Nayagam, B. A. & Richardson, R. T. Hair cell regeneration after ATOH1 gene therapy in the cochlea of profoundly deaf adult guinea pigs. PLoS One 9, e102077 (2014).
Mizutari, K. et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 77, 58–69 (2013).
Liu, Z. et al. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J Neurosci 32, 6600–6610 (2012).
Liu, Z., Fang, J., Dearman, J., Zhang, L. & Zuo, J. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic Atoh1 expression. PLoS One 9, e89377 (2014).
Romand, R. & Chardin, S. Effects of growth factors on the hair cells after ototoxic treatment of the neonatal mammalian cochlea in vitro. Brain Res 825, 46–58 (1999).
Sun, H., Lin, C. H. & Smith, M. E. Growth hormone promotes hair cell regeneration in the zebrafish (Danio rerio) inner ear following acoustic trauma. PLoS One 6, e28372 (2011).
Pan, N. et al. A novel Atoh1 “self-terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS One 7, e30358 (2012).
Yang, S. M. et al. Regeneration of stereocilia of hair cells by forced Atoh1 expression in the adult mammalian cochlea. PLoS One 7, e46355 (2012).
Cox, B. C., Liu, Z., Lagarde, M. M. & Zuo, J. Conditional gene expression in the mouse inner ear using Cre-loxP. J Assoc Res Otolaryngol 13, 295–322 (2012).
Mellado Lagarde, M. M. et al. Selective ablation of pillar and deiters’ cells severely affects cochlear postnatal development and hearing in mice. J Neurosci 33, 1564–1576 (2013).
Arnold, K. et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329 (2011).
Kiernan, A. E. et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434, 1031–1035 (2005).
Shi, F. et al. beta-Catenin is required for hair-cell differentiation in the cochlea. J Neurosci 34, 6470–6479 (2014).
Hume, C. R., Bratt, D. L. & Oesterle, E. C. Expression of LHX3 and SOX2 during mouse inner ear development. Gene Exp Patterns: GEP 7, 798–807 (2007).
Smeti, I., Watabe, I., Savary, E., Fontbonne, A. & Zine, A. HMGA2, the architectural transcription factor high mobility group, is expressed in the developing and mature mouse cochlea. PLoS One 9, e88757 (2014).
Dabdoub, A. et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA 105, 18396–18401 (2008).
Cai, T. et al. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the atoh1 transcription factor. J Neurosci 35, 5870–5883 (2015).
Scheffer, D. I., Shen, J., Corey, D. P. & Chen, Z. Y. Gene Expression by Mouse Inner Ear Hair Cells during Development. J Neurosci 35, 6366–6380 (2015).
Wang, T. et al. Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in damaged neonatal mouse utricle. Nature Comms 6, 6613 (2015).
Maass, J. C. et al. Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Frontiers Cellular Neurosci 9, 110 (2015).
Laos, M. et al. DNA damage signaling regulates age-dependent proliferative capacity of quiescent inner ear supporting cells. Aging 6, 496–510 (2014).
Chen, Y. et al. Cotransfection of Pax2 and Math1 promote in situ cochlear hair cell regeneration after neomycin insult. Sci Rep 3, 2996 (2013).
Korrapati, S., Roux, I., Glowatzki, E. & Doetzlhofer, A. Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS One 8, e73276 (2013).
Walters, B. J. et al. Auditory hair cell-specific deletion of p27Kip1 in postnatal mice promotes cell-autonomous generation of new hair cells and normal hearing. J Neurosci 34, 15751–15763 (2014).
Kelly, M. C., Chang, Q., Pan, A., Lin, X. & Chen, P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci 32, 6699–6710 (2012).
Liu, Z., Owen, T., Fang, J., Srinivasan, R. S. & Zuo, J. In vivo Notch reactivation in differentiating cochlear hair cells induces Sox2 and Prox1 expression but does not disrupt hair cell maturation. Dev Dyn 241, 684–696 (2012).
Liu, Z. et al. Regulation of p27Kip1 by Sox2 maintains quiescence of inner pillar cells in the murine auditory sensory epithelium. J Neurosci 32, 10530–10540 (2012).
Shi, F., Hu, L. & Edge, A. S. Generation of hair cells in neonatal mice by beta-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci USA 110, 13851–13856 (2013).
Liu, Z., Walters, B. J., Owen, T., Kopan, R. & Zuo, J. In vivo visualization of Notch1 proteolysis reveals the heterogeneity of Notch1 signaling activity in the mouse cochlea. PLoS One 8, e64903 (2013).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Mak, A. C., Szeto, I. Y., Fritzsch, B. & Cheah, K. S. Differential and overlapping expression pattern of SOX2 and SOX9 in inner ear development. Gene Exp Patterns: GEP 9, 444–453 (2009).
Jacques, B. E. et al. A dual function for canonical Wnt/beta-catenin signaling in the developing mammalian cochlea. Development 139, 4395–4404 (2012).
Ono, K. et al. FGFR1-Frs2/3 signalling maintains sensory progenitors during inner ear hair cell formation. PLoS Genet 10, e1004118 (2014).
Acknowledgements
This research was supported in part by funding from the National Institutes of Health (grants DC006471 (J.Z.), 1R21DC013879 (J.Z.), P30CA21765 (St. Jude)), American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital, the Office of Naval Research (grants N000140911014, N000141210191, N000141210775 (J.Z.)), the National Organization for Hearing Research (NOHR) Foundation (B.W.), the Hearing Health Foundation (Emerging Research Grant, B.W.) and The Hartwell Foundation (Individual Biomedical Research Award, J.Z.).
Author information
Authors and Affiliations
Contributions
B.J.W., T.Y. and J.Z. contributed to the experimental design. B.J.W. and T.Y. performed experiments, B.J.W., T.Y. and J.Z. analyzed data, B.J.W., T.Y. and J.Z. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Walters, B., Yamashita, T. & Zuo, J. Sox2-CreER mice are useful for fate mapping of mature, but not neonatal, cochlear supporting cells in hair cell regeneration studies. Sci Rep 5, 11621 (2015). https://doi.org/10.1038/srep11621
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep11621
This article is cited by
-
Abnormal Innervation, Demyelination, and Degeneration of Spiral Ganglion Neurons as Well as Disruption of Heminodes are Involved in the Onset of Deafness in Cx26 Null Mice
Neuroscience Bulletin (2024)
-
Deletion of the Notch ligand Jagged1 during cochlear maturation leads to inner hair cell defects and hearing loss
Cell Death & Disease (2022)
-
GATA3 maintains the quiescent state of cochlear supporting cells by regulating p27kip1
Scientific Reports (2021)
-
Gfi1Cre mice have early onset progressive hearing loss and induce recombination in numerous inner ear non-hair cells
Scientific Reports (2017)
-
SOX2 is required for inner ear neurogenesis
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.