Abstract
Asthma is a chronic inflammatory disease. Around 5 to 10% of patients classified as having severe asthma can-not be adequately controlled despite the use of all currently available therapeutic approaches. Previous studies have revealed the potential important role of miRNAs in the regulation of a variety of inflammatory processes, including asthma. Expression of selected miRNAs, specifically let-7a, miR-21 and miR-223, that were shown to have important roles in asthma pathogenesis, were analyzed in bronchial biopsies of 24 patients with asthma, 12 mild and 12 severe and 10 controls with no chronic disease. We found significantly reduced expression of let-7a in bronchial biopsies from patients with severe asthma in comparison to patients with mild asthma as well as in comparison to the non-asthmatic controls. On the other hand, no significant differences in miR-21 and miR-223 expression were found between the different groups analyzed. Reduced let-7a levels in bronchial biopsies of patients with severe therapy-resistant asthma could not only be used as a potential biomarker to discriminate between different asthma phenotypes, but also might be a target for modulation of treatment at the inflammatory site for a group of patients that are most affected and still lack effective treatment.
Similar content being viewed by others
Introduction
Asthma is a chronic inflammatory disease with exaggerated bronchoconstriction after provocation with specific or non-specific stimuli. It is one of the most common chronic diseases, affecting more than 300 million people of all ages worldwide. The incidence continues to increase and thus represents a serious challenge for public health system1,2. Therapy currently primarily consists of inhaled corticosteroids, long- and short-acting beta-agonists and leukotriene antagonists, which are not directed at the underlying disease, but rather to symptom relief1,2,3,4. A majority of patients achieve good symptom control and minimal exacerbation using regular controller therapy; however, up to 10% of patients with severe asthma can-not be adequately controlled despite maximal therapy1,2,5. The variability in treatment response is based on large asthma clinical heterogeneity and understanding the pattern of airway inflammation and molecular factors regulating those processes could be helpful in defining asthma endotypes and understanding why therapy is not effective in all patients6.
MicroRNAs (miRNAs), 19–25 nucleotides long non-coding small RNAs involved in regulation of gene expression through a process known as RNA interference7,8,9, may play a role in orchestrating the phenotypic programming of immune and airway epithelial cells to enhance the production of cytokines and other mediators that result in the inflammation that characterizes asthma3,4,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25. Therefore, miRNAs may contribute to asthma pathology. Distinct miRNA expression was found in samples from subjects with asthma, such as epithelial brushings or circulating T cells12,26 and experiments have also shown altered expression in sensitized and challenged mouse asthma models27. Numerous miRNAs have been found to be dysregulated in asthma; however, the replication level is relatively low and data on miRNA expression in human lung tissue are still scarce3,4,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25. MiRNAs have been found to play critical roles in regulating key pathogenic mechanisms in inflammation, including polarization of adaptive immune responses and activation of T cells (e.g., miR-21), regulation of eosinophil development (e.g., miR-21 and miR-223)4 and regulation of IL-13, a key cytokine in allergic lung inflammation (e.g., let-7a)11.
Many miRNAs identified as either tumor suppressors or oncogenes in lung cancer were also reported to be involved in the immune response; for example, the let-7 family, the most abundant miRNAs in the lungs28. The let-7 family is involved in pro-inflammatory cytokine production29 because it inhibits IL-13 expression and represents a major regulatory mechanism for modulating IL-13 secretion and thereby TH2 inflammation, characteristic for TH2 high asthma30. There was reduced expression of let-7a in samples from subjects with asthma12,15 and overexpression of let-7a reduced airway inflammation and hyper-responsiveness in the lungs of asthma mouse models10,11.
Among the top miRNAs expressed in inflamed tissue is miR-214. MiR-21 is thought to play an important role in immunity, in maintaining the effector phase of T cells31 and in regulation of TH2 immune responses through IL-12 targeting32. In asthma, miR-21 was found to be up-regulated32 and its expression was associated with eosinophilic TH2 high asthma33.
MiR-223 was found to be one of the candidates involved in asthma pathogenesis when asthmatic mouse lung tissue miRNA expression was profiled using a microarray27. This miRNA is involved in granulocytopoiesis and granulocyte activation and is thought to be a regulator for preventing hyper-inflammatory states34. It was found to be down-regulated in human asthmatic T cells26 and in mice elevated pro-inflammatory cytokines IL-6 and IL-1β were found as a result of miR-223 down-regulation upon induction with LPS35. In humans, IL-6 levels are known to be elevated in asthma and also correlate with lung function and IL-13 levels36.
MiRNAs let-7a, miR-21 and miR-223 were shown to either directly or indirectly repress the translation of several crucial factors with well-defined roles in asthma pathogenesis, such as signal transducer and activator of transcription 3 (STAT3), interleukins (IL-6, IL-1β, IL-13), interferon gamma (IFN-γ), transforming growth factor, beta receptor (TGF-β receptor), toll-like receptor 4 (TLR4) and vascular endothelial growth factor (VEGF)3,4,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25. Dysregulated miRNAs could have a role in the pathogenesis of asthma and could be used as potential biomarkers to discriminate between different asthma phenotypes. Furthermore, miRNAs are promising targets for a novel and effective therapeutic strategy for modulation of inflammatory response for a group of patients that still lack effective treatment.
Let-7a, miR-21 and miR-223 seem to be involved in modulation of inflammation in the lungs and therefore we hypothesized that they are differentially expressed in bronchial biopsies of patients with severe asthma.
Results
Patient characteristics
The characteristics of the study groups from which the lung biopsies were derived are shown in Table 1. Patients with severe asthma had significantly reduced FEV1% predicted and Tiffeneau index (TI) in comparison to patients with mild asthma and non-asthmatic controls (P < .01 and P < .001, respectively). Vital capacity was lower in the severe asthma group than in the control group (P < .05). In addition, patients with severe asthma experienced more asthma exacerbations per year (P < .001) and also required oral corticosteroids more often (P = .003, OR 33.0, 95%CI 2.9−374.5) in comparison to those with mild asthma. Other clinical characteristics did not differ between the study groups.
miRNA expression
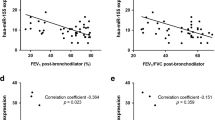
Expression of selected miRNAs was determined in bronchial biopsy tissue samples and normalized to a sample from the control group. When we stratified asthma patients according to severity, we found significantly reduced expression of let-7a in bronchial biopsies from patients with severe asthma in comparison to bronchial tissues from patients with mild asthma as well as to bronchial tissues from the control group (Figure 1). When analyzing the entire group of asthma patients (24), we did not observe any difference in expression of let-7a between patients and controls. Moreover, no significant differences in miR-21 and miR-223 expression were found between the different groups analyzed (Figure 2).
Discussion
Asthma is a complex disorder of the immune system characterized by chronic inflammation, airflow obstruction and bronchial hyper-responsiveness. Tightly balanced pathways, networks of activators and suppressors are needed for proper regulation of immune responses. MiRNAs have been shown to be important regulators of variety of immunologically driven processes3,4,21,22,23 and are emerging as important biomarkers in the pathogenesis of asthma3,4,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25. However, the data on miRNA expression in the lungs of human subjects with asthma are still scarce. Several miRNAs, such as let-7a, miR-21 and miR-223, have been shown to have a role in airway inflammation, TH1/TH2 polarization and eosinophil development either directly or indirectly repress the translation of several crucial factors and were shown to be differentially expressed in asthma3,4,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25.
In the present study we analyzed the expression of three miRNAs (let-7a, miR-21 and miR-223) in bronchial biopsy samples from patients with mild and severe asthma and non-asthmatic controls. The expression of let-7a was significantly reduced in severe asthma compared to mild asthma and controls, whereas the expressions of miR-21 and miR-223 were not altered.
Altered expression of let-7a has been found in many diseases; for example, in various cancers, Alzheimer's disease and immune system diseases, such as asthma, allergies, allergic rhinitis and atopic dermatitis37. Let-7a is highly expressed in human lungs19 and it is hypothesized to have an anti-inflammatory role38. In mouse asthma models with overexpressed let-7a, reduction of airway inflammation and hyper-responsiveness was observed11 and it has numerous targets related to asthma pathogenesis, such as IL-13, ADRB2, TLR4 and TGF-ß receptor15,38,39. It is hypothesized that the main contribution of let-7a in asthma is targeting of IL-13 and IL-6 and thus modulating TH2 responses in lung inflammatory processes, which was shown on mouse models and cell cultures3,10,11,16. Increased TH2 response and airway inflammation in asthma is often associated with severe therapy-resistant asthma, characterized by frequent and severe exacerbations, poor asthma control and severe airflow obstruction30,40. Moreover, a correlation was found between IL-6 and lung function5,36, which further supports the potential role of down-regulated let-7a in severe asthma, for which poor lung function is typical1,5,6.
Regarding the let-7a expression in asthma, inconsistent results have been found because a few studies have indicated no difference between healthy subjects and asthma patients19,25. Williams et al. found no differences in miRNA expression in patients with mild asthma in comparison to healthy controls19, whereas a study by Jardim et al. found differentially expressed miRNAs in patients with mild asthma, but let-7a expression did not differ between patients with mild asthma and controls25. On the other hand, other studies have demonstrated reduced let-7a expression in asthma patients. Pinkerton et al. examined miRNA expression in exhaled breath condensate in asthma patients and found let-7a levels to be lower than in COPD and in healthy controls14. A microarray analysis performed on bronchoalveolar lavage fluid exosomes of asthma patients revealed 8 qPCR confirmed altered miRNAs, including down-regulated let-7a and miR-21. Pathway analysis of differentially expressed miRNAs belonged to JAK-STAT signaling, cytokine network and cytokine and inflammatory response13. Let-7a was also found to be reduced in the airway epithelial cells12 and serum15 of asthma patients. Thus, reduced expression of let-7a in bronchial biopsies from patients with severe asthma is consistent with the findings of other studies3,10,11,12,13,14,15,19,25.
Similar to let-7a, inconsistent results on miR-21 and miR-223 expression in asthma have also been reported12,15,19,26,27. MiR-21 was significantly up-regulated in asthma regardless of treatment33 and its expression was also altered in various asthma mouse models32; however, as in ours, no changes were found in other studies19. MiR-223 was previously shown to be down-regulated in asthma26; however, we found no alternation in its expression. MiR-21 and miR-223 are thought to play a role in eosinophil development4 and they were both up-regulated in eosinophilic esophagitis patients41. Various complex immunological processes that simultaneously occur in the lungs of asthma patients could be the reason for inconsistent results between studies3. Some miRNA expressions are constitutively altered in disease, whereas others need an induction in order to be expressed differentially42.
In addition to the miRNAs analyzed in our study, several others were shown to be involved in asthma. For example, miR-146a is involved in TLR and cytokine signaling4,23 and miR-29, miR-126 and miR-155 play important roles in airway remodeling4,20. In animal models, modulation of several miRNAs, specifically let-7a, miR-21, miR-145, miR-126 and miR-106a, resulted in decreased asthma severity and inflammation3,4,13,14,19. Therefore, miRNAs were not only shown to affect several mechanisms in asthma, but also show potential utility as novel targets for therapy3,4,10,21,22,23.
Depending on asthma severity, different doses of therapy are required and this raises a question about therapy influence on miRNA expression in our study. Reduced let-7a and non-altered miR-21 and miR-223 could be due to therapy; however, Williams et al. demonstrated that different treatment regimens and different doses of inhaled corticosteroids do not affect miRNA expression19. In addition to the relatively small sample size, one concern regarding our study design might be related to the appropriate choice of controls. Our study investigated miRNA expression in bronchial biopsies and as controls we included 10 patients in whom bronchoscopy was indicated as part of the diagnostic procedure. In all controls, chronic lung disease was excluded by various routine tests. Six were diagnosed as GERD and four of them as hemoptysis with normal radiologic, endoscopic and lung function findings. Even though the controls used in our study were not healthy and both conditions might be potentially associated with lung inflammation, we believe these controls represent a valid control group for our experiments. MiRNA expression did not vary significantly between different controls, specifically between patients with GERD and hemoptysis. Furthermore, Smith et al. analyzed miRNA expression in GERD patients and found that none of the miRNA analyzed in our study were differentially expressed43. Even though the results of our preliminary study are promising, additional prospective studies and larger sample sizes are needed to improve our knowledge of asthma pathogenesis and to elucidate the mechanisms that lead to the development of this inflammatory disease. This will eventually lead to new insights for the development of targeted therapies for more specific and effective treatment.
In summary, our results show that let-7a is down-regulated in patients with severe asthma. These results could contribute to understanding the severe asthma mechanism and let-7a could potentially be used as a biomarker to discriminate between different asthma phenotypes. Most importantly, let-7a might be a promising target for novel therapeutic approaches for a group of patients that can-not be adequately controlled despite the use of all currently available therapeutic approaches1,2,3,4,5,6, especially because it was shown that inhibiting let-7a could tone down asthma symptoms10. Many studies already performed in this field, as well as a number of preclinical and clinical studies, predict exponential growth of RNA-interference–based methods as possible new target therapies to regulate gene expression. Nevertheless, RNA-interference–based gene silencing approaches have proven to be successful in treating several disorders, including retinal degeneration, dominantly inherited brain and skin diseases, cancer and metabolic disorders44.
Methods
Patients
Twenty-four patients treated at the University Clinic for Respiratory and Allergic Diseases Golnik from 2010 to 2012 with a diagnosis of asthma according to Global Initiative for Asthma (GINA) guidelines1 were included. We divided them into two subgroups according to the asthma severity (GINA guidelines). Twelve patients were classified as having mild asthma with controlled asthma on low to moderate doses of inhaled corticosteroids (ICS). The other 12 patients had severe asthma, used high doses of ICS and had frequent exacerbations that required systemic corticosteroids. They were in a stable phase of the disease, with no evidence of exacerbation in the past 4 weeks. As controls we used 10 patients with no known chronic disease; in six of them, bronchoscopy was indicated because of prolonged cough that was finally attributed as a consequence of gastro-esophageal reflux disease (GERD) and four of them had hemoptysis with normal radiologic, endoscopic and lung function findings. The characteristics of the study groups are shown in Table 1.
Biopsy Procedures
Bronchial biopsies were taken during diagnostic procedures with a flexible bronchoscope and were immediately formalin-fixed and then paraffin-embedded using standard procedures. All patients were in a stable phase of the disease, with no evidence of exacerbation in the past 4 weeks. This study was conducted in accordance with the amended Declaration of Helsinki. All subjects gave written informed consent and the study was approved by the Slovenian National Medical Ethics Committee (approval number 95/06/13).
Selection of specific miRNAs involved in asthma pathogenesis
To select miRNAs involved in asthma pathogenesis, available publications3,4,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25 and several target prediction databases were searched, including miRanda (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl), TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRbase (http://microrna.sanger.ac.uk/). The selection criteria were: miRNAs that were previously shown to be expressed in the lungs and miRNAs associated with asthma and/or asthma-related genes/proteins that were reported in at least two published studies and/or target prediction databases. Based on these findings, three miRNAs involved in the pathway strongly associated with asthma, specifically let-7a, miR-21 and miR-223, were selected. Selected miRNAs were shown to have roles in asthma pathogenesis, either directly or indirectly repressing the translation of crucial factors such as STAT3, IL-6, IL-1β, IL-13, IFN-γ, TGF-β receptor, TLR4 and VEGF.
RNA extraction, reverse transcription and real-time PCR
Total RNA was extracted from 10 FFPE tissue sections 5 µm thick using the miRNeasy FFPE Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's instructions. Quantitative PCR was used to analyze the expression of selected miRNAs as previously described45. Isolated RNA was reverse transcribed using miRNA-specific RT primers (RNU6B, hsa-let-7a, hsa-miR-21 and hsa-miR-223) and a TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, California, USA). RT products were used in a real-time PCR reaction with the TaqMan miRNA assays (Applied Biosystems) and TaqMan Fast Advanced Master Mix (Applied Biosystems) according to the manufacturer's instructions. All samples were run in triplicate. The RNU6B was used as an endogenous control for normalization of the target miRNAs and a sample from the control group was used as a calibrator. Relative expression was calculated using the ΔΔCt method. The fold change was determined by 2−ΔΔCt, where ΔCt = (average of triplicate CtTarget miRNA − average triplicate CtRNU6B) and ΔΔCt = (ΔCtSample − ΔCt Sample from control group). Real-time PCR was performed on an ABI PRISM 7500 Real-Time PCR System (Sequence Detection System instrument equipped with SDS version v2.0.5 software; Applied Biosystems).
Statistical Analyses
The distribution of data was determined using the D'Agostino and Pearson omnibus normality test. The strength of association between miRNA expression levels and other clinical variables was analyzed with the Mann–Whitney U-test, unpaired t-test, or Fisher's exact test, as appropriate. Statistical analyses were performed using GraphPad Prism 5 software (San Diego, California, USA) and probability values of P < .05 were accepted as significant.
References
Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention. Global Initiative for Asthma website. http://www.ginasthma.org/guidelines-gina-report-global-strategyfor-asthma.html. Updated 2012. Accessed July 15, 2013.
Weiss, S. T. New approaches to personalized medicine for asthma: Where are we? J. Allergy Clin. Immunol. 129, 327–334 (2009).
Rebane, A. & Akdis, C. A. MicroRNAs: Essential players in the regulation of inflammation. J. Allergy Clin. Immunol. 132, 15–26 (2013).
Lu, T. X. & Rothenberg, M. Diagnostic, functional and therapeutic roles of microRNA in allergic diseases. J. Allergy Clin. Immunol. 132, 3–13 (2013).
O'Byrne, P. M. et al. The poorly explored impact of uncontrolled asthma. Chest 143, 511–523 (2013).
Lemanske, R. F. Jr. & Busse, W. W. Asthma: clinical expression and molecular mechanisms. J. Allergy Clin. Immunol. 125, S95–102 (2010).
Eulalio, A., Huntzinger, E., Nishihara, T., Rehwinkel, J., Fauser, M. & Izaurralde, E. Deadenylation is a widespread effect of miRNA regulation. RNA 15, 21–32 (2009).
Friedman, R. C., Farh, K. K., Burge, C. B. & Bartel, D. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Rana, T. M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell. Biol. 8, 23–36 (2007).
Polikepahad, S. et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J. Biol. Chem. 285, 30139–30149 (2010).
Kumar, M. et al. let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 128, 1077–1085 (2011).
Solberg, O. D. et al. Airway epithelial miRNA expression is altered in asthma. Am. J. Respir. Crit. Care Med. 186, 965–974 (2012).
Levänen, B. et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 131, 894–903 (2013).
Pinkerton, M. et al. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease and healthy adults. J. Allergy Clin. Immunol. 132, 217–219 (2013).
Panganiban, R. P. L., Pinkerton, M. H., Maru, S. Y., Jefferson, S. J., Roff, A. N. & Ishmael, F. T. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am. J. Clin. Exp. Immunol. 1, 154–165 (2012).
Iliopoulos, D., Hirsch, H. A. & Struhl, K. An epigenetic switch involving NF-κB, Lin28, let-7 microRNA and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009).
Lu, T. X. et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization and the severity of delayed-type hypersensitivity. J. Immunol. 187, 3362–3373 (2011).
Sharma, A. et al. Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. J. Appl. Physiol. (1985) 113, 459–464 (2012).
Williams, A. E. et al. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS One 4, e5889 (2009).
Nakano, T. et al. Lower levels of hsa-mir-15a, which decreases VEGFA, in the CD4(+) T cells of pediatric patients with asthma. J. Allergy Clin. Immunol. 132, 1224–1227 (2013).
Omran, A., Elimam, D. & Yin, F. MicroRNAs: new insights into chronic childhood diseases. Biomed. Res. Int. 2013, 291826 (2013).
Sessa, R. & Hata, A. Role of microRNAs in lung development and pulmonary diseases. Pulm. Circ. 3, 315–328 (2013).
Wang, J. W., Li, K., Hellermann, G., Lockey, R. F., Mohapatra, S. & Mohapatra, S. Regulating the Regulators: microRNA and Asthma. World Allergy Organ. J. 4, 94–103 (2011).
Collison, A., Herbert, C., Siegle, J. S., Mattes, J., Foster, P. S. & Kumar, R. K. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm. Med. 11, 29 (2011).
Jardim, M. J., Dailey, L., Silbajoris, R. & Diaz-Sanchez, D. Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. Am. J. Respir. Cell. Mol. Biol. 47, 536–542 (2012).
Seumois, G. et al. An integrated nano-scale approach to profile miRNAs in limited clinical samples. Am. J. Clin. Exp. Immunol. 1, 70–89 (2012).
Garbacki, N. et al. MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS One 6, e16509 (2011).
Takamizawa, J. et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64, 3753–3756 (2004).
Oglesby, I. K., McElvaney, N. G. & Greene, C. M. MicroRNAs in inflammatory lung disease--master regulators or target practice? Respir. Res. 11, 148 (2010).
Peters, J. B. et al. Health status measurement in patients with severe asthma. Respir. Med. 108, 278–286 (2014).
Wu, H. et al. Mir-21 is thought to play an important role in immunity, in maintaining effector phase of T cells. PLoS One 2, e1020 (2007).
Lu, T. X., Munitz, A. & Rothenberg, M. E. J. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. Immunol 182, 4994–5002 (2009).
Wu, X. B. et al. Overexpression of microRNA-21 and microRNA-126 in the patients of bronchial asthma. Int. J. Clin. Exp. Med. 7, 1307–1312 (2014).
Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. MicroRNAs modulate hematopoietic lineage differentiation. Science 6, 83–86 (2004).
Chen, Q. et al. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS One 7, e42971 (2012).
Neveu, W. A. et al. Elevation of IL-6 in the allergic asthmatic airway is independent of inflammation but associates with loss of central airway function. Respir. Res. 11, 28 (2010).
Russo, F. et al. miRandola: Extracellular Circulating microRNAs Database. PLoS ONE 7, e47786 (2012).
Gosh, B. Involvement of microRNA in Asthma: New perspective in respiratory biology. Indian J. Allergy Asthma Immunol. 27, 3–8 (2013).
Wang, X. & Naqa, M. E. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 24, 325–332 (2008).
Wenzel, S. E. Asthma phenotypes: the evolution from clinical to molecular approaches. Nature Medicine 18, 716–725 (2012).
Lu, T. X. et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids and assessment as disease biomarkers. J. Allergy Clin. Immunol. 129, 1064–1075 (2012).
Ha, T. Y. MicroRNAs in Human Diseases: From Cancer to Cardiovascular Disease. Immune Netw. 11, 135–154 (2011).
Smith, C. M. et al. Impact of gastro-oesophageal reflux on microRNA expression, location and function. BMC Gastroenterol. 13, 4 (2013).
Davidson, B. L. & McCray, P. B. Jr. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 12, 329–340 (2011).
Jusufović, E. et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non-small-cell lung cancer. PLoS ONE 7, e45577 (2012).
Acknowledgements
The authors are grateful to all the patients that participated in the study. This work was supported by the Slovenian Research Agency (grant no. P3-0360).
Author information
Authors and Affiliations
Contributions
M.R., P.K., M.Ž. and M.M.M. contributed to designing the study, M.M.M. contributed to recruiting subjects, performing the bronchial biopsies and collecting relevant data, M.R. and M.Ž. performed the expression experiments, M.R., M.M.M. and P.K. carried out the data analysis and data interpretation, I.K. contributed to performing the immunohistochemical staining and collecting relevant clinical data and M.R., M.Ž. and M.M.M. wrote the main manuscript. All authors reviewed and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Rijavec, M., Korošec, P., Žavbi, M. et al. Let-7a is differentially expressed in bronchial biopsies of patients with severe asthma. Sci Rep 4, 6103 (2014). https://doi.org/10.1038/srep06103
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06103
This article is cited by
-
MiR-223 plays a protecting role in neutrophilic asthmatic mice through the inhibition of NLRP3 inflammasome
Respiratory Research (2020)
-
A network-based approach to uncover microRNA-mediated disease comorbidities and potential pathobiological implications
npj Systems Biology and Applications (2019)
-
A plasmonic colorimetric strategy for visual miRNA detection based on hybridization chain reaction
Scientific Reports (2016)
-
Long-term efficacy and safety of omalizumab in patients with persistent uncontrolled allergic asthma: a systematic review and meta-analysis
Scientific Reports (2015)
-
The Expression and Significance of the Plasma Let-7 Family in Anti-N-methyl-d-aspartate Receptor Encephalitis
Journal of Molecular Neuroscience (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.