Abstract
Influenza A viruses cause the respiratory illness influenza, which can be mild to fatal depending on the strain and host immune response. The flu polymerase acidic (PA), polymerase basic 1 (PB1) and polymerase basic 2 (PB2) proteins comprise the RNA-dependent RNA polymerase complex responsible for viral genome replication. The first crystal structures of the C-terminal domain of PA (PA-CTD) in the absence of PB1-derived peptides show a number of structural changes relative to the previously reported PB1-peptide bound structures. The human A/WSN/1933 (H1N1) and avian A/Anhui1/2013 (H7N9) strain PA-CTD proteins exhibit the same global topology as other strains in the absence of PB1, but differ extensively in the PB1 binding pocket including a widening of the binding groove and the unfolding of a β-turn. Both PA-CTD proteins exhibited a significant increase in thermal stability in the presence of either a PB1-derived peptide or a previously reported inhibitor in differential scanning fluorimetry assays. These structural changes demonstrate plasticity in the PA-PB1 binding interface which may be exploited in the development of novel therapeutics.
Similar content being viewed by others
Introduction
Influenza A and B viruses cause the infectious disease influenza, commonly referred to as flu, in the respiratory tract of mammals and birds. The current standard of care is a seasonal vaccine that is typically effective for only a subset of the host population and ineffective against the more virulent strains of influenza that can cause pandemics. These vaccines target viral surface proteins which mutate quickly, hence the need for yearly vaccinations. Therefore, there is a significant unmet medical need for the development of novel therapeutics against influenza viruses. The influenza viral RNA-dependent RNA polymerase (RdRp) complex is highly conserved and may be a suitable target for therapeutics that are effective across many viral strains. The Influenza A virus (IAV) RdRp consists of three separate polypeptide subunits: the polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2) and the polymerase acidic protein (PA)1. The IAV RdRp associates with the viral RNA genome and the viral nucleoprotein (NP) to form the viral ribonucleoprotein (vRNP) complex responsible for transcription and translation2. PB1 contains the nucleic acid polymerase catalytic subunit3. Although the IAV RdRp likely uses the same two metal ion mechanism common to all polymerases4 and contains the common GD(D/N) divalent metal ion binding catalytic motif at residues 304–306, the remaining primary sequence may be unlike any other polymerase of known structure. PB1 forms the core of the IAV RdRp complex, interacting with PA through its highly conserved N-terminus5 and PB2 through its C-terminus. PA contains an N-terminal endonuclease domain (PAN) that interacts with PB26 and a C-terminal domain (PA-CTD) that interacts with PB1 (Figure 1). An mRNA frameshift results in a distinctively different protein, termed PA-X, which represses cellular gene expression and thereby modulates infectivity and the host immune response7. Crystal structures have been determined for PAN as apo8,9, in complex with nucleotides10 or inhibitors11,12 and for PA-CTD in complex with small peptides derived from the N-terminus of the PB1 protein13,14. A structure of the PB1-PB2 interface has also been determined15. No crystal structures have yet been determined for full length PA, the PA-PB1-PB2 complex, or PA-X.
Schematic representation of full length IAV PA.
The Influenza A virus (IAV) polymerase acidic (PA) protein contains an N-terminal endonuclease domain (PAN) as well as a C-terminal PB1-binding domain (PA-CTD). The PA-X protein arises due to a -1 nucleotide frameshift in the open reading frame7. A structure of PAN is shown in blue ribbons representation with Mn2+ ions depicted as orange spheres and inhibitor DPBA shown in sticks12. A structure of the PA-CTD is shown in gray with a peptide derived from the N-terminus of the PB1 protein in red14. The chemical structure of a previously reported inhibitor19 which was also used in this study is also shown.
Viral polymerases have been successful therapeutic targets for other single-stranded RNA viruses such as HIV and Hepatits C. In addition to inhibitors of the PAN enzyme11,12,16, there have been reports of peptide inhibitors17,18 and small molecule inhibitors19,20,21 that bind to PA and disrupt the PA-PB1 interaction. Computational docking and other structure-based drug design strategies could exploit the bound state of PA-CTD with PB1-peptide removed13,14. Structural information on the apo state of the PA-CTD could impact such activities by providing a state of the protein prior to binding PB1 and thus be useful in the development of compounds that target this protein-protein interaction. Here we present the first crystal structures of PA-CTD in the absence of PB1-derived peptides, showing that although the global protein topology appears the same, there is significant movement of portions of the PB1 binding site. The protein constructs used here appear to be functionally active in that they can be shown to bind to PB1-derived peptides as well as an inhibitor reported previously in the literature. These results improve our understanding of the structural features of the influenza PA protein and how they may be exploited for the development of novel therapeutics.
Results
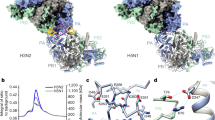
We cloned full length IAV PA (residues 1–716) and PA-CTD (residues 254–716) from the 1933 Wilson-Smith human H1N1 strain and the 2013 Anhui avian H7N9 strain using codon engineered synthetic genes22,23 and overexpressed these proteins as N-terminal hexahistidine-Smt fusion proteins (see Methods). Although we were unable to express full length H1N1 PA, we generated purified H1N1 PA-CTD as well as both full length and PA-CTD H7N9 protein samples in quantities (multiple mg) necessary to support crystallographic analysis. These proteins appear folded, possessing a single melting transition at temperatures of 43°C for either H7N9 full length or PA-CTD and 38°C for H1N1 PA-CTD, as determined by differential scanning fluorimetry24 assays (Figure 2). Full length 1933 H1N1 has previously been reported to bind to a peptide corresponding to the first 15 amino acids of the PB1 protein with an IC50 of 41 nM18. In the presence of a peptide derived from the first 25 amino acids of the PB1 protein, we observed a significant thermal stabilization of all three PA proteins, with single transition melting temperatures of 56°C (+13°C ΔTm) for full length H7N9 PA, 57°C (+14°C ΔTm) for H7N9 PA-CTD and 54°C (+16°C ΔTm) for H1N1 PA-CTD (Figure 2). Furthermore, the thermal stabilization observed upon binding to PB1-derived peptide suggests each protein sequence tested is functionally active in that they are capable of binding a PB1-derived peptide. Such large thermal stabilizations are not surprising given the large hydrophobic surface area buried at the PA/PB1 interface in the PA/PB1-peptide crystal structures13,14. We also investigated binding of our PA protein samples with an inhibitor (Figure 1) that targets the PB1 binding site on PA with an IC50 of 20–30 μM in either ELISA or FluA minireplicon assays19. Due to solubility challenges with the compound at 300 μM concentration, we were unable to obtain a single melting transition, but did observe two-peak melting transitions which resemble a mixture of the apo peak transition and the PB1-derived peptide bound transition (Figure 2). Thus, these protein samples may be capable of binding small molecule inhibitors in addition to binding PB1-derived peptides.
Differential scanning fluorimetry analysis of IAV PA.
(a), Analysis of full length influenza A virus 2013 avian H7N9 polymerase acidic (PA-FL) by differential scanning fluorimetry (DSF) as apo (red trace), in the presence of a PB1-derived peptide (orange) and with a previously reported fluorinated inhibitor (blue). (b), DSF analysis of the PA-CTD from 2013 avian H7N9 as apo (red), in the presence of a PB1-derived peptide (orange) and with a previously reported fluorinated inhibitor (blue). (c), DSF analysis of the PA-CTD from 1933 human H1N1 as apo (red), in the presence of a PB1-derived peptide (orange) and a previously reported fluorinated inhibitor (blue).
We determined crystal structures of the IAV PA-CTD in the absence of PB1-derived peptides at 1.9 Å resolution for H1N1 and at 2.2 Å resolution for H7N9 (Figure 3 and Table 1). The crystals obtained for these two proteins possess the same hexagonal crystal form (Table 1) and thus exhibit many similar features in the final structures. However, differences in the c-axis result in a number of residues that are disordered and solvent exposed in the H7N9 structure but are ordered and involved in crystal packing in the H1N1 structure. The overall structures of PA-CTD (residues 254–716) from the Wilson-Smith 1933 H1N1 human strain and the Anhui 2013 H7N9 avian strain are similar to previously reported structures of PA-CTD in complex with PB1-derived peptides from either avian H5N113 or human H1N114, but with clear differences in two peripheral loops and the PB1 binding site (Figure 3c). The Cα backbone r.m.s.d. values for apo H1N1 compared with PB1-bound H1N1 (PDB ID 2ZNL14) and PB1-bound H5N1 (PDB ID 3CM813) were 1.18 Å (377 similar residues) and 1.03 Å (380 similar residues), respectively. The Cα backbone r.m.s.d. values for apo H7N9 compared with PB1-bound H1N1 (PDB ID 2ZNL14) and PB1-bound H5N1 (PDB ID 3CM813) were 0.98 Å (355 similar residues) and 0.86 Å (368 similar residues), respectively. The Cα backbone r.m.s.d. value for apo H1N1 and apo H7N9 is 0.53 Å (375 similar residues). The W422-E436 loop connecting α4 and α5 distal to the PB1-binding site is well ordered in Wilson-Smith 1933, while largely disordered in both the 2013 Anhui avian H7N9 and the 1997 Hong Kong avian H5N113 and adopts a different conformation in Puerto Rico 1934 human H1N114. This region contains a single A432V point variant in the 1933 H1N1 apo structure that is involved in crystal packing. Taken as a whole, the two apo structures and two PB1 peptide-bound structures show a high degree of structural variability in the W422-E436 loop, indicative of flexibility. The loop connecting α3 and α4 that spans residues N373 to P398 was mostly ordered in the avian H5N1 structure but was disordered in the other three structures, preventing further analysis.
Comparison of crystal structures of PA-CTD in the absence or presence of PB1-derived peptides.
(a), Crystal structure of apo 1933 Wilson-Smith human H1N1 PA-CTD solved at 1.9 Å resolution. (b), Crystal structure of apo 2013 Anhui avian H7N9 PA-CTD solved at 2.2 Å resolution. (c), Global overlay of apo 1933 Wilson-Smith human H1N1 PA-CTD (green), apo 2013 Anhui avian H7N9 (blue) and 1998 Hong Kong avian H5N1 PA-CTD13 (gray) in complex with a peptide derived from the N-terminus of the PB1 protein (red). (d), Close up of the overlay shown in panel c highlighting differences in active site residues along α8, α9 and the β8–β9 turn. (e), Overlay of the apo and PB1-derived peptide-bound structures as in panels (c) and (d) highlighting residues at the base of the peptide binding groove which were predicted to bind to small molecule inhibitor via in silico docking experiments (Q408, N412, K643, W706)20,21 which shows little conformational variability between the apo and peptide-bound structures as well as a second predicted binding site (L666, Q670, R673, F710) toward the top of the cavity which shows considerably more variability between the apo and PB1-peptide bound structures.
Of particular interest are structural differences that occur at the PB1 binding site (Figure 3). Out of the 36 PA-CTD residues which contact the N-terminus of the PB1-derived peptide, only three (N409S, I621V and E630D) exhibit sequence differences between the strains studied here and a number of historic pandemic strains (Figure 4). In the PB1 peptide-bound structures13,14, α4 and β8 contact the β-strand of the PB1 peptide, while β8-β9, α10 and α11-α13 contact the α-helix of the PB1 peptide. Without the PB1 peptide, both α11-α13 and α5-α8-α9 move outward to increase the size of this cavity by ca. 5 Å at its widest point (Figure 3d). Virtually all of these residues exhibit higher than average B-factors indicating relatively high thermal motion in the absence of the PB1 peptide. In addition, the β8-β9 turn is largely disordered; in the H1N1 apo structure the C-terminal residues of β9 form an alternative conformation, packing against a crystal lattice contact in the apo H1N1 structure. Interestingly, the residues remaining from this stretch and residual electron density point 180° away from orientation of the β8-β9 turn observed in the PB1 bound structure. P625 and K626 form the turn between β8 and β9 in the PB1-peptide bound structure, but in the apo state these residues have moved 24 and 19 Å, respectively, from their positioning in the PB-1 bound state. These results strongly suggest that this turn is highly mobile in the absence of PB1. Without the PB1 peptide present in the complex, M595 of α8, F611 of α9 and I633, V636 and L640 of α10 are exposed (Figure 3d). In the PB1 peptide-bound state, M595, F611 and I633 coordinate W619 of β8 whereas V636 and L640 contact L8 of PB1 α1. However, none of these residues are truly solvent exposed in the H1N1 apo crystal structure, but rather are involved in crystal packing with the loop that spans β6–β7 of a symmetry related molecule. Differences in packing result in these residues being significantly more solvent exposed in the apo H7N9 structure.
Sequence analysis of virulent avian and human influenza strains.
(a), Multiple sequence alignment comparing the PA-CTD sequences from 1933 Wilson-Smith human H1N1 and 2013 Anhui avian H7N9 strains solved here with several highly virulent and pandemic strains. (b), Mapping the sequence differences between the two structures solved here and the pandemic strains onto the 1933 H1N1 apo structure with residue 552 highlighted in red. (c), The β6–β7 turn which includes the T552S mutation which allows bypass of the species-species restriction.
Discussion
The two apo crystal structures of human 1933 H1N1 and avian 2013 H7N9 presented here expand our understanding of the structural basis for influenza virulence, PB1 binding and potential interactions with small molecules that target the PA/PB1 interface. The Wilson-Smith 1933 H1N1 strain was the first flu virus cultured in the laboratory and has been well studied, due to its high sequence homology with the 1918 pandemic Spanish flu believed to be responsible for nearly 50 million deaths (Figure 4a). Typically, avian flu cannot replicate in human cells; however, it was shown that acquisition of a human PA subunit or simply the T552S mutation may allow avian flu strains to bypass the species-species restriction of the avian polymerase complex25. In the 1934 H1N1 human strain structure, the loop containing residue S552 was disordered14; our 1933 H1N1 human flu PA structure now permits detailed analysis of this residue critical for virulence (Figure 4b). In the human strain, the side chain of S552 forms hydrogen bonds with the backbone nitrogen of I554 and the carbonyl of G555 in the solvent exposed β6–β7 turn (Figure 4c). In the avian H5N1 PA-CTD PB1 peptide complex13, T552 is modeled in a different rotamer conformation with the side chain hydroxyl pointing toward a solvent channel; however, in our avian H7N9 apo structure T552 adopts the same conformation as in our human H1N1 apo structure and here, rather, the side chain methyl point toward solvent. Further investigation into the position of this critical residue in the presence of larger binding partners will be necessary to determine how PA-CTD with T552 may prevent full assembly or function of the RdRp in human IAV.
A number of small molecule inhibitors which target the PA/PB1 interface have been identified by virtual screening experiments19,20,21. Many of these compounds were predicted to bind at the base of the PB1 binding cleft and interact with Q408, N412, K643 and W70620,21, which are involved in recognition of V3-N4-P5-V6 of the PB1-derived peptide13,14. Residues Q408, N412 and W706 as well as nearby hydrophobic residues F411 and C415 exhibited the same conformation across the two apo and the two PB1 peptide-bound crystal structures (Figure 3e). K643 adopts a slightly different rotamer conformation in the absence of the PB1 peptide. All six of these residues are solvent exposed in our apo H1N1 and H7N9 PA-CTD crystal structures. The major difference in this region is the lack of the β8–β9 turn, which could possibly cap inhibitor binding from the opposite face as the Q408-N412-W706 motif, similar to its interaction with the PB1 peptide. The second hot spot for ligand binding identified by in silico docking experiments correspond to the binding site for the C-terminal residues of the PB1-derived peptide (L10-P13 of PB1), including residues L666, Q670 and R673 on α11 and F710 on α13 of PA. The widening of this region in the absence of the PB1 peptide results in differences in positioning of several of these residues. Thus, the base of the PB1 binding cleft (residues Q408, N412 and W706) appears largely unchanged between the apo and PB1 peptide-bound states and may provide a platform for initial interactions with potential ligands. Other elements such as the β8–β9 turn and α11 appear mobile and may provide conformational malleability within the binding site. Thus, it may be possible to exploit both the rigid platform at the base of the PB1 binding cleft and the more flexible elements on the periphery in the design and development of therapeutics which abrogate the PA-PB1 protein-protein interaction.
Though the IAV viral polymerase is highly conserved between strains, the structural differences highlighted here reveal the need to examine homologous PA-PB1 complexes in the development of therapeutics using a structure-based approach. In addition to examining the more rigid and highly conserved areas of the binding pocket, less conserved residues may present challenges in compound design and could potentially lead to varied binding affinities between strains. Thus, careful consideration should be taken when selecting strains for developing novel therapeutics.
Methods
Overexpression and purification
PA C-terminal domains from both the 1933 Wilson-Smith IAV H1N1 (A/WS/1933/H1N1 UniProt ID P15659) and the 2013 Anhui IAV H7N9 (A/Anhui1/2013/H7N9 UniProt ID S5MT15) strains spanning residues 254 through the native C-terminus (716) were overexpressed in E. coli as a His-Smt fusion using protocols similar to that which resulted in several PB2 crystal structures26,27,28. Full length (1–716) PA from the 2013 Anhui IAV H7N9 strain was expressed in the same manner. Briefly, the proteins were purified by nickel affinity chromatography followed by cleavage and removal of the fusion tag with ULP-1 protease and a subtractive nickel column. The final samples were obtained after sizing with a Sephacryl S-100 SEC column in 25 mM Tris pH 8.0, 200 mM NaCl, 1% v/v glycerol and 1 mM TCEP.
Differential scanning fluorimetry (DSF) analysis
Differential scanning fluorimetry24 experiments were performed on the IAV PA-CTD from H1N1 as well as the full length PA and PA-CTD from H7N9 using Sypro Orange dye (Sigma-Aldrich) on an Opticon Monitor II from 20 to 100°C at 1 mg/mL protein (12–19 μM). The use of 100 mM sodium acetate pH 6.2 resulted in improved curve profiles (Figure 2) relative to the original purification buffer (25 mM Tris pH 8.0, 200 mM NaCl, 1% v/v glycerol and 1 mM TCEP). The PB1-derived peptide (NH2-MDVNPTLLFLKVPVQNAISTTFPYTKK) was obtained as a custom peptide synthesis at >90% purity from GenScript and included two lysine residues at the C-terminus to improve solubility. The previously identified PA inhibitor19 N-[3-(aminocarbonyl)-5,6-dihydro-4H-cyclopenta[b]thien-2-yl]-7-(difluoromethyl)-5-phenylpyrazolo[1,5-a]pyrimidine-3-carboxamide was obtained from ChemBridge (Catalog No. 7914306). The peptide or inhibitor were used at 200 and 300 μM concentrations, respectively. All samples contained 5% v/v DMSO for consistency and all samples were run in triplicate with the average temperature reported. The standard deviations were <1°C.
Crystallization
Crystals of human H1N1 PA-CTD (SSGCID target ID InvaN.07057.a) grew from the Morpheus screen29 condition G5 (10% w/v PEG 8000, 20% v/v ethylene glycol, 0.1 M MOPS/HEPES pH 7.5 and 20 mM of each sodium formate, ammonium acetate, trisodium citrate, sodium/potassium D/L tartrate and sodium oxamate) at 20.45 mg/mL (384 μM). Crystals of avian H7N9 PA-CTD (SSGCID target ID InvaQ.07057.a) grew from the PACT screen condition A10 (200 mM MgCl2, 0.1 M sodium acetate pH 5.0, 20% PEG 6000) at 15 mg/mL (282 μM). Crystals were grown using the sitting drop vapor diffusion method at 289 K with 0.4 μL protein and 0.4 μL precipitant equilibrated against 80 μL of reservoir.
X-ray data collection and structure determination
For X-ray diffraction data collection, crystals of H1N1 PA-CTD were vitrified directly, whereas crystals of H7N9 PA-CTD were harvested into a cryo-protectant containing precipitant supplemented with 20% v/v ethylene glycol. X-ray data (Table 1) were collected at the APS LS-CAT and reduced with XDS30 using a single crystal for each target. The H1N1 PA-CTD structure was solved by molecular replacement in Phaser31 using PDB ID 2ZNL14 as the search model and the H7N9 PA-CTD structure was solved using the apo 1933 H1N1 PA-CTD structure. The final H1N1 PA-CTD model (Table 1) was obtained after iterative refinement in Refmac532, manual model building in Coot33 and validation in MolProbity34. The final H7N9 PA-CTD was obtained after iterative refinement in both Refmac532 and Phenix35 with manual model building in Coot33 and validation in MolProbity34.
Additional information
Accession codes: Atomic coordinates for the reported structures have been deposited with the Protein Data Bank under accession codes 4IUJ and 4P9A.
References
Resa-Infante, P., Jorba, N., Coloma, R. & Ortin, J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8, 207–215 (2011).
Nagata, K., Kawaguchi, A. & Naito, T. Host factors for replication and transcription of the influenza virus genome. Rev. Med. Virol. 18, 247–260 (2008).
Sivasubramanian, N. & Nayak, D. P. Sequence analysis of the polymerase 1 gene and the secondary structure prediction of polymerase 1 protein of human influenza virus A/WSN/33. J. Virol. 44, 321–329 (1982).
Steitz, T. A. A mechanism for all polymerases. Nature 391, 231–232 (1998).
Binh, N. T., Wakai, C., Kawaguchi, A. & Nagata, K. The N-terminal region of influenza virus polymerase PB1 adjacent to the PA binding site is involved in replication but not transcription of the viral genome. Frontiers in Microbiol. 4, 398 (2013).
Hemerka, J. N. et al. Detection and characterization of influenza A virus PA-PB2 interaction through a bimolecular fluorescence complementation assay. J. Virol. 83, 3944–3955 (2009).
Shi, M. et al. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J. Virol. 86, 12411–12413 (2012).
Dias, A. et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918 (2009).
Yuan, P. et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458, 909–913 (2009).
Zhao, C. et al. Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center. J. Virol. 83, 9024–9030 (2009).
Bauman, J. D. et al. Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors. ACS Chem. Biol. 8, 2501–2508 (2013).
Kowalinski, E. et al. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 8, e1002831 (2012).
He, X. et al. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454, 1123–1126 (2008).
Obayashi, E. et al. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454, 1127–1131 (2008).
Sugiyama, K. et al. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28, 1803–1811 (2009).
Chen, E. et al. Computation-guided discovery of influenza endonuclease inhibitors. ACS Med. Chem. Lett. 5, 61–64 (2014).
Chase, G., Wunderlich, K., Reuther, P. & Schwemmle, M. Identification of influenza virus inhibitors which disrupt of viral polymerase protein-protein interactions. Methods 55, 188–191 (2011).
Wunderlich, K. et al. Identification of high-affinity PB1-derived peptides with enhanced affinity to the PA protein of influenza A virus polymerase. Antimicrob. Agents Chemother. 55, 696–702 (2011).
Muratore, G. et al. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc. Natl. Acad. Sci. USA 109, 6247–6252 (2012).
Pagano, M. et al. The fight against the influenza A virus H1N1: synthesis, molecular modeling and biological evaluation of benzofurazan derivatives as viral RNA polymerase inhibitors. ChemMedChem 9, 129–150 (2014).
Tintori, C. et al. High-throughput docking for the identification of new influenza A virus polymerase inhibitors targeting the PA-PB1 protein-protein interaction. Bioorg. Med. Chem. Lett. 24, 280–282 (2014).
Lorimer, D. et al. Gene composer: database software for protein construct design, codon engineering and gene synthesis. BMC Biotechnol. 9, 36 (2009).
Raymond, A. et al. Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer. BMC Biotechnol. 9, 37 (2009).
Niesen, F. H., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protoc. 2, 2212–2221 (2007).
Mehle, A., Dugan, V. G., Taubenberger, J. K. & Doudna, J. A. Reassortment and mutation of the avian influenza virus polymerase PA subunit overcome species barriers. J. Virol. 86, 1750–1757 (2012).
Armour, B. L. et al. Multi-target parallel processing approach for gene-to-structure determination of the influenza polymerase PB2 subunit. J. Vis. Exp. June 26, 76; 10.3791/4225 (2013).
Smith, E. R. et al. The Protein Maker: an automated system for high-throughput parallel purification. Acta Cryst. F 67, 1015–1021 (2011).
Yamada, S. et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 6, e1001034 (2010).
Gorrec, F. The MORPHEUS protein crystallization screen. J Appl. Crystallogr. 42, 1035–1042 (2009).
Kabsch, W. Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Cryst. D 60, 2126–2132 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst. D 66, 12–21 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D 66, 213–221 (2010).
Acknowledgements
The authors thank the entire SSGCID team. This research was funded by the National Institute of Allergy and Infectious Diseases, National Institute of Health, Department of Health and Human Services, under Federal Contract number HHSN272201200025C and HHSN272200700057C. The expression plasmids and surplus protein samples can be obtained from either the contact authors or through www.ssgcid.org and the raw X-ray diffraction data can be obtained through www.csgid.org.
Author information
Authors and Affiliations
Contributions
S.O.M., D.W.B., I.P., B.L.S., R.S., P.J.M., D.L. and T.E.E. conceived, designed and supervised the project. G.H., K.M. and V.T. nominated the targets to the SSGCID pipeline and suggested domain boundaries. J.B., J.K., S.R.B. and A.C.R. performed cloning, expression and protein purification. R.O.B. performed DSF experiments. J.A., J.W.F. and T.E.E. performed crystallization experiments, collected the X-ray data and solved the crystal structures. S.O.M., J.A., J.W.F., R.O.B. and T.E.E. analyzed the data. S.O.M., J.A., R.O.B., I.P. and T.E.E. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Moen, S., Abendroth, J., Fairman, J. et al. Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1. Sci Rep 4, 5944 (2014). https://doi.org/10.1038/srep05944
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05944
This article is cited by
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
-
Characteristic amino acid changes of influenza A(H1N1)pdm09 virus PA protein enhance A(H7N9) viral polymerase activity
Virus Genes (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.