Abstract
Model organisms containing deletion or mutation in a glycosyltransferase-gene exhibit various physiological abnormalities, suggesting that specific glycan motifs on certain proteins play important roles in vivo. Identification of the target proteins of glycosyltransferase isozymes is the key to understand the roles of glycans. Here, we demonstrated the proteome-scale identification of the target proteins specific for a glycosyltransferase isozyme, β1,4-galactosyltransferase-I (β4GalT-I). Although β4GalT-I is the most characterized glycosyltransferase, its distinctive contribution to β1,4-galactosylation has been hardly described so far. We identified a large number of candidates for the target proteins specific to β4GalT-I by comparative analysis of β4GalT-I-deleted and wild-type mice using the LC/MS-based technique with the isotope-coded glycosylation site-specific tagging (IGOT) of lectin-captured N-glycopeptides. Our approach to identify the target proteins in a proteome-scale offers common features and trends in the target proteins, which facilitate understanding of the mechanism that controls assembly of a particular glycan motif on specific proteins.
Similar content being viewed by others
Introduction
It has become obvious that the post-translational modification of proteins plays a critical role in the regulation of biological events. Protein glycosylation is one of the most common post-translational modifications in eukaryotes. The glycan moiety was found to be functionally linked to a wide variety of cellular events, including cell adhesion, immune reaction and signal transduction1,2. Thus, one of the most central and challenging issues in glycobiology is to elucidate the role of glycans. Glycan biosynthesis is not a template-based process and dictated by the concerted reactions of multiple glycosyltransferases with different substrate specificities3. More than 170 glycosyltransferases and sulfotransferases have been cloned so far (GlycoGene Database in Japan Consortium for Glycobiology and Glycotechnology DataBase; JCGG-DB, http://riodb.ibase.aist.go.jp/rcmg/ggdb).
One of the most promising approaches for elucidating the physiological role of a glycan is to disrupt the glycosyltransferase gene responsible for the biosynthesis of that particular glycan motif. Model organisms carrying deletion or mutation in the glycosyltransferase gene indeed exhibit various physiological abnormalities4,5,6. Subsequent studies strongly suggested that the loss of specific glycan motifs on certain proteins influences functions of the proteins and leads to physiological abnormalities7,8,9. Thus, to determine proteins responsible for the abnormalities and also their molecular mechanisms, it is highly desirable to identify the target proteins for the glycosyltransferase responsible for the biosynthesis of the glycan motif.
The aim of our study is the identification of the target proteins for glycosyltransferases in a proteome-scale. Here, as an initial case, we attempted to identify the target proteins for β4GalT-I, the most characterized glycosyltransferase that was cloned first in the history of glycobiology10,11. Out of seven members of the β4GalT subfamily (β4GalT-I through β4GalT-VII), at least five isozymes (β4GalT-I, -II, -III, -IV and -V) are able to transfer the Gal residue to the terminal GlcNAc via a β1,4-linkage and form N-acetyl lactosamine (Galβ1,4GlcNAc; LacNAc)12, which is one of the most widely expressed terminal motifs of N- and O-glycans on vertebrate proteins3. Varieties of terminal glycan motifs, such as (sialyl) Lewis X and HNK-1, are formed on LacNAc. In addition, a recent quantitative PCR analysis demonstrated the expression of multiple β4GalT genes in various mouse organs13. Consistent with this observation, the finding of residual β4GalT activity in some organs, such as the liver and brain, of β4GalT-I−/− mice suggested that β4GalTs other than β4GalT-I are also expressed in these organs14,15. We have shown in vitro that the isozymes of β4GalT (β4GalT-I to β4GalT-V) exhibited different branch-specificities for N-glycan16. Thus, to elucidate their distinct contributions to β1,4-galactosylation in vivo, as well as to determine the role of Galβ1,4-terminated glycans, it is essential to identify the target proteins for each β4GalT isozyme. Among all the isozymes, β4GalT-I is believed to be the major β4GalT isozyme responsible for the biosynthesis of biologically important glycan motifs. In fact, several studies using β4GalT-I−/− mice suggested that the glycans synthesized by β4GalT-I are related in epithelial cell growth and differentiation15, inflammatory response17, skin wound healing18 and IgA nephropathy development19. Another group reported that these glycans are also involved in the anterior pituitary hormone function and fertilization14.
Until recently, there had been no suitable technology available to identify the target proteins of a particular glycosyltransferase isozyme in a proteome-scale. Presence and co-expression of other isozymes interfere with the identification of the target proteins of the isozyme. The fact that isozymes, just like β4GalTs, could produce one particular glycan motif under the physiological condition complicates the situation. Schjoldager et al. addressed this issue by utilizing their “SimpleCell” system20,21. Using the system, they identified the target proteins specific for an isozyme of polypeptide GalNAc transferase in the HepG2 cell line. A family of polypeptide GalNAc transferases initiates protein O-glycosylation by transferring an α-GalNAc residue to Ser/Thr residue(s). Their technique is systematic and highly suited for identification of the α-GalNAc attachment sites of O-glycans. It is now also required to establish an approach for determination of the target proteins belonging to other glycosyltransferase families which, in particular, involved in biosynthesis of N-glycans.
While the glycoproteomic approach, coupling the LC/MS shotgun protein identification method with the lectin-affinity purification method, is a powerful mean to identify a large number of N-glycosylated proteins with a specific glycan motif, it is theoretically impossible to distinguish which isozyme is actually responsible for producing the glycan motif on each protein. Using this approach, Zielinska et al. have identified over 2,000 N-glycosylated proteins in various mouse tissues22. For example, in the mouse liver, approximately 500 glycoproteins were considered to carry Galβ1,4-terminated N-glycans because those proteins were identified by lectin-affinity purification using Ricinus communis agglutinin 120 (RCA120), a plant lectin with specific affinity for Galβ1,4-terminated glycans (Lectin Frontier DataBase in JCGG-DB; http://riodb.ibase.aist.go.jp/rcmg/glycodb/LectinSearch and previous reports23,24). However, it was impossible to distinguish which of these proteins were targets for which isozyme of β4GalT.
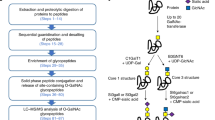
We have developed a comprehensive identification method of N-glycosylated proteins with a particular glycan motif, called “lectin-IGOT-LC/MS method”, by combining the lectin-mediated glycopeptide-capture and LC/MS-based identification of the core peptides25,26,27. In the present study, we applied this lectin-IGOT-LC/MS method for the comparative glycoproteomic analysis of wild-type (WT) and β4GalT-I−/− mice. Accordingly, the target proteins specific for β4GalT-I were identified by comparing the proteins that were glycosylated with Galβ1,4-terminated glycans (“proteins carrying Galβ1,4-terminated glycans”) in WT and β4GalT-I−/− mice (Fig. 1), as the β4GalT-I-specific target proteins are not expected to be β1,4-galactosylated in β4GalT-I−/− mice. We utilized the RCA120-IGOT-LC/MS method to determine proteins carrying Galβ1,4-terminated glycans in the liver and by comparing the lists of these glycoproteins in the WT and β4GalT-I−/− mice, we identified attractive candidates for the β4GalT-I-specific target proteins on a proteome-scale while multiple β4GalT isozymes were present.
Schematic representation of methods described in this study.
To identify β4GalT-I-specific target proteins, we determined and compared presence of Galβ1,4-terminated glycans on individual liver proteins of WT and β4GalT-I−/− mice. We used the RCA120-IGOT-LC/MS method to comprehensively identify proteins carrying Galβ1,4-terminated glycans. The glycoproteins identified from the WT mice were target proteins of β4GalT-I and other co-expressed β4GalT(s), whereas the glycoproteins identified from the β4GalT-I−/− mice were target proteins of β4GalTs other than β4GalT-I. Thus, those glycoproteins that were present in the WT mice, but not in the β4GalT-I−/− mice, were regarded as the β4GalT-I-specific target proteins, because the β4GalT-I-specific target proteins should not be β1,4-galactosylated in the β4GalT-I−/− mice.
Results
Large-scale identification of proteins carrying Galβ1,4-terminated glycans in WT and β4GalT-I−/− mouse liver using the RCA120-IGOT-LC/MS method
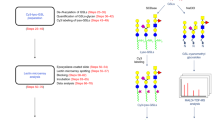
First, we prepared a subset of glycopeptides carrying Galβ1,4-terminated glycan from protease digests of the liver extracts from WT and β4GalT-I−/− mice using an RCA120-immobilized column (Fig. 2a). In this approach, selective isolation of glycopeptides carrying Galβ1,4-terminated glycans is a critical step for identifying proteins carrying the desired glycan motif with high reliability. The purity of the glycopeptide samples further assures that the identified glycoproteins are not false positives, but are the true target proteins of the isozyme. We found that the Galβ1,4-terminated and Galβ1,3-terminated glycans exhibited significantly different retention times on the RCA120-immobilized column (Supplementary Fig. S1). This result suggested that the RCA120-immobilized column is reliable for specific isolation of glycopeptides carrying Galβ1,4-terminated glycans. Together with previously published results, it was suggested that RCA120 binds to a Galβ1,4-terminated glycan more preferentially than to a Galβ1,3-terminated glycan (Lectin Frontier DataBase in JCGG-DB and previous reports23,24). We, therefore, washed the RCA120-immobilized column exhaustively (until A230 of the washing <0.005) to ensure complete removal of glycopeptides carrying Galβ1,3-terminated glycans as well as all nonspecifically bound (glyco)peptides. We then eluted the bound glycopeptides from the column using lactose as the competing sugar.
Flowchart outlining the steps involved in identifying the β4GalT-I-specifc target proteins using the RCA120-IGOT-LC/MS method and summary of results.
(a) Outline of steps used for specific isolation of glycopeptides carrying Galβ1,4-terminated glycans and subsequent identification of glycoproteins carrying Galβ1,4-terminated glycans. First, we prepared a subset of glycopeptides carrying Galβ1,4-terminated glycans from the protease digested liver extracts of the WT and β4GalT-I−/− mice by using the RCA120-immobilized column. After the purification of glycopeptides by HILIC, the peptides were labeled with 18O by the IGOT reaction as described25. Each labeled peptide was subsequently analyzed by the LTQ/Orbitrap Velos nanoLC-MS/MS and the core peptide of each glycopeptide was identified by searching a mouse protein database using the Mascot algorithm. Numbers in the diagram indicate the number of identified glycoproteins that carry Galβ1,4-terminated glycans. (b) Plot showing number of glycoproteins identified from six individual shotgun LC/MS analyses of each sample. Results obtained from two biological replicates of each genotype are shown. (c) Summary of β4GalT-I-specifc target proteins identified by the RCA120-IGOT-LC/MS method. Candidate target proteins were determined by comparing the list of identified glycoproteins of the WT and β4GalT-I−/− mice. As a result, 1,007 glycoproteins, each carrying Galβ1,4-terminated glycans, were identified from the WT mice and they were categorized into two categories: 181 proteins as “candidate target proteins of β4GalT-I” (indicated by the shaded area) and 826 proteins as “sharable target proteins”, which are assumed to be targets of more than one β4GalT isozyme, including β4GalT-I.
Subsequently, to remove any contaminating non-glycopeptide, we performed hydrophilic interaction liquid chromatography (HILIC) using a TSKgel Amide-80 column28. After purification, N-glycosylated Asn residues of the glycopeptides were specifically labeled with 18O by using peptide N-glycanase-F (PNGase-F)25,26,27. Then, the proteins carrying Galβ1,4-terminated glycans in both WT and β4GalT-I−/− mice were identified by shotgun LC-MS/MS analysis.
Shotgun identification of proteins carrying Galβ1,4-terminated glycans
The effectiveness of our approach is largely influenced by the coverage of concerned glycoproteome during shotgun LC-MS/MS analysis. To increase the coverage of proteins carrying Galβ1,4-terminated glycans, we performed repeated scans on a sample (WT or β4GalT-I−/− mouse) using an LC/MS coupled to LTQ/Orbitrap Velos mass spectrometer. Subsequent processing of the data generated by the LC-MS/MS analysis by the Mascot algorithm revealed a large number of proteins carrying Galβ1,4-terminated glycans from both WT and β4GalT-I−/− samples. Based on the selection criteria described in Methods, we selected “identified peptide”. An “identified peptide” was then accepted as an “N-glycopeptide" when it contained one or more 18O-labeled Asp residue(s) in a consensus tripeptide sequence for N-glycosylation (Asn-Xaa-[Ser/Thr/Cys], where Xaa is any amino acid except proline). Detailed results on peptide identification are summarized in Supplementary Tables S1 and S2. It is noteworthy that after the HILIC purification step, among all the identified peptides, 75.8% (1,672/2,221) were accepted as N-glycopeptides, which suggested that our peptide enrichment procedure is highly specific and N-glycopeptide identification results are reliable. After the sixth round of scanning and data processing, the number of identified glycoproteins was saturated (Fig. 2b), suggesting that our list covers a substantial portion of proteins carrying Galβ1,4-terminated glycans. Consequently, from 12 rounds of analysis for each genotype, 1,391 18O-labeled unique peptides identified from the WT liver were assigned to 1,007 proteins and 1,259 18O-labeled unique peptides identified from the β4GalT-I−/− liver were assigned to 995 proteins. Overall, a total of 1,176 proteins were identified as glycoproteins carrying Galβ1,4-terminated glycans. The false positive rate for peptide identification using the criteria described in Methods was estimated by a decoy database approach for every Mascot database search. The average false positive rate in 12 rounds of analysis was 0.80% for the WT samples and 0.87% for the β4GalT-I−/− samples.
Subsequently, we compared the lists of identified glycoproteins from the WT and β4GalT-I−/− mice to determine which proteins were exclusively identified in the WT mice. There was a relatively large overlap (70%; 826/1,176) between the proteins carrying Galβ1,4-terminated glycans identified from both WT and β4GalT-I−/− mice, except for 181 proteins which were found only in the WT mice (Fig. 2c). These 181 proteins were regarded as the candidates for the target proteins specific to β4GalT-I.
Molecular characterization of the candidates for the β4GalT target proteins
To elucidate the regulation mechanism of β1,4-galactosylation and the contribution of each β4GalT isozyme to the regulation process in vivo, we compared the molecular properties of the candidate target proteins identified in this study. For simplicity, we assumed that the proteins identified in both the WT and β4GalT-I−/− mice are “sharable” target proteins, because they could be β1,4-galactosylated by β4GalT-I and other β4GalT isozymes.
Our identified protein lists included several sets of homologous proteins that shared the same peptide match (e.g., isoforms, splicing variants, family proteins). To minimize redundancy caused by the existence of homologous proteins, we selected proteins identified by a peptide(s) shown in bold face type in the Mascot database-search result as the representative of these homologous proteins. (“Bold” means a peptide match to a query appearing in the report for the first time.) Then, to reduce the false positive identification of β4GalT-I-specific target proteins resulting from the chance detection during the shotgun LC/MS analysis, only proteins that were identified in 3 or more rounds of analysis (out of 12 separate rounds of analysis) were taken into consideration. As a result, 70 proteins were selected as the plausible candidates for the β4GalT-I-specific target proteins to be considered in this study. Subsequently, we determined the molecular characteristics of these candidate target proteins and 611 proteins of the sharable target proteins. The list of subjected proteins in this study is shown in Supplementary Table S3.
Since β4GalTs are tethered to the Golgi membrane and the enzyme reaction for β1,4-galactosylation predominantly occurs in the Golgi lumen, we next focused on the localization of the target proteins in the Golgi apparatus. We used web-based protein analysis tools, SignalP29, TMHMM30 and SOSUI31 to predict whether the candidate target proteins contain any signal peptide and/or transmembrane (TM) segment. Subsequently, we categorized each protein as a soluble-type or membrane-type protein. Among the plausible candidates for the β4GalT-I-specific target proteins, 44.3% (31/70) were predicted to be soluble-type proteins, while 24.9% (152/611) of the sharable target proteins were in the same category (Fig. 3a and also detailed in Supplementary Table S3). Thus, β4GalT-I, as compared to other β4GalT isozymes, shows a tendency to play a dominant role in β1,4-galactosylation of secretory proteins, rather than that of membrane proteins.
Predicted membrane topology and classification of the candidate target proteins.
(a) The candidate target proteins were classified as soluble-type, membrane-type or ambiguous, based on signal peptide and TM prediction analyses results. Out of the 753 proteins, 189 proteins were predicted as “soluble-type”, 428 proteins were predicted as “membrane-type” and the remaining 136 proteins were termed as “ambiguous”. Distributions of “soluble-type” and “membrane-type” target proteins in each target protein category are indicated. A value in parentheses indicates the number of identified glycoproteins in each group. (b) Number of TM domains in the membrane-type target proteins as predicted by the TMHMM algorithm. The TMHMM algorithm was used to predict the number of TM domains. The average number of predicted TM domains found among the target proteins was 2.5 for the candidates for the β4GalT-I-specific target proteins, 4.5 for the sharable target proteins and 5.5 for the KO-only proteins. A value in parentheses indicates the number of identified glycoproteins in each category.
The programs used here for predicting the TM segment can also predict the number of TM domains within the protein. The frequency for finding membrane-type proteins with a single TM domain among the candidates for the β4GalT-I-specific target proteins was significantly higher than the sharable target proteins: 76.9% vs 42.3% by using the TMHMM algorithm (Fig. 3b). In contrast, out of 163 putative membrane proteins with more than five TM domains, only 2 proteins were found in the candidates for the β4GalT-I-specific target proteins (see details in Supplementary Table S3). More than 90% of the candidates for the β4GalT-I-specific target proteins were predicted to possess relatively low number (one to three) of TM domains. Consistent with these observations, the average calculated number of TM domains was 2.5 for the candidates for the β4GalT-I-specific target proteins and 4.5 for the sharable target proteins. Similar results were also obtained when the SOSUI algorithm was used for the TM domain prediction analysis (Supplementary Fig. S2). These results suggest that the β1,4-galactosylation of membrane proteins containing a multiple number of TM domains is not solely controlled by β4GalT-I, but other β4GalT isozymes also play dominant roles.
Next, we focused on the molecular size of the candidate target proteins. Molecular weight (MW) of each protein was calculated from its amino acid sequence without taking any protein modification into consideration. Although the average MW of the candidates for the β4GalT-I-specific target proteins (105.2 kDa) was higher than that of the sharable target proteins (82.9 kDa), no remarkable difference was found in their overall MW distributions (Fig. 4a). Remarkably, putative membrane proteins with predicted MW over 200 kDa were frequently found in the candidates for the β4GalT-I-specific target proteins (Fig. 4b). Thus, 23% (6/26) of the putative membrane proteins in the candidates for the β4GalT-I-specific target proteins exhibited MW over 200 kDa, while only 3.4% (12/352) of the putative membrane proteins in the sharable target protein category were found in the same MW range. Among the putative membrane proteins, the average MW of the candidates for the β4GalT-I-specific target proteins (140.8 kDa) was significantly higher than that of the sharable target proteins (80.1 kDa). These results suggested that β4GalT-I predominantly contributes to the β1,4-galactosylation of membrane proteins, especially those with higher MW. This was, however, not observed in the putative soluble target proteins (Fig. 4c). Thus, we could assume that the membrane proteins that are target proteins of β4GalT-I exhibited distinctive molecular features, namely high MW and single TM domain.
Molecular size distribution of the candidate target proteins.
(a) Molecular size distribution of the candidate target proteins (irrespective to their predicted localization). MW was calculated from the amino acid sequence of each target protein without taking any protein modification into consideration. Average MW was 105.2 kDa for the candidates for the β4GalT-I-specific target proteins, 82.9 kDa for the sharable target proteins and 59.6 kDa for the KO-only proteins. (b) Molecular size distribution of the membrane-type target proteins. Average MW was 140.8 kDa for the candidates for the β4GalT-I-specific target proteins, 80.1 kDa for the sharable target proteins and 64.2 kDa for the KO-only proteins. (c) Molecular size distribution of the soluble-type target proteins. Average MW was 88.7 kDa for the candidates for the β4GalT-I-specific target proteins, 95.4 kDa for the sharable target proteins and 56.2 kDa for the KO-only proteins.
Finally, we examined the molecular features of proteins that were solely identified in the β4GalT-I−/− mice (“KO-only proteins”). Of the 72 proteins categorized as KO-only proteins (Supplementary Table S3) following criteria similar to those used for selecting the plausible candidates for the β4GalT-I-specific target proteins, more membrane-type proteins were found than soluble-type proteins (Fig. 3a). Nonetheless, the predicted number of TM domains (Fig. 3b) and the MW distribution profiles (Fig. 4a–c) of the KO-only proteins were similar to those of the sharable target proteins. We have provided a possible explanation for finding these proteins only in the β4GalT-I−/− mice in Discussion.
Collectively, characterization of such a large number of the candidate target proteins allowed us to suggest that β4GalTs catalyze β1,4-galactosylation of a wide variety of proteins irrespective to their molecular sizes, functions, subcellular localization, or membrane topology. On the other hand, the candidates for the β4GalT-I-specific target proteins exhibited several distinctive characteristics: (1) soluble proteins, rather than membrane proteins, are more frequently found in this group and (2) membrane proteins found in this group usually contain single TM domain and exhibit high MW (>200 kDa). Our results also suggest that the membrane topology and molecular size of a protein might influence whether the protein could be a target for a specific β4GalT in vivo.
Discussion
To the best of our knowledge, this is the first report describing common features and trends in the target proteins of a particular glycosyltransferase isozyme through a glycoproteomic approach. The unique information on the molecular characteristics was obtained owing to the deep coverage of the concerned glycoproteome and the large number of the candidate target proteins, which reinforce reliability of our approach. Molecular characterization of the candidate target proteins using the bioinformatic tools offers novel and unique cues to understand the control mechanism for assembly of a particular glycan motif on certain proteins. It has been hardly understood how glycan structures attached on individual proteins are controlled in vivo. Intriguingly, our results raised possibility, which is quite novel, that β1,4-galactosylation of a specific protein population is predominantly controlled by β4GalT-I and not so much by other β4GalT-isozymes. No other previous study has shown such distinctive contribution of a β4GalT isozyme in vivo and thus we proved that our approach can provide unique information which could not be obtained using other methods currently available. It should be emphasized here that our bioinformatic analyses provided attractive information, but the molecular characteristics above mentioned are still speculative and further examinations in future studies are necessary.
β1,4-galactosylation of proteins is known to decrease in β4GalT-I−/− mice15. Thus, one might expect a significantly less number of identified proteins in β4GalT-I−/− mice than in WT mice. Contrary to this expectation, however, the decreased β1,4-galactosylation hardly had any effect on the number of identified proteins in the β4GalT-I−/− mouse liver (Fig. 2b,c). We believe that this is not due to a high false-positive rate of the identification from the β4GalT-I−/− mice, as we could specifically isolate glycopeptides carrying Galβ1,4-terminated glycans using the RCA120-immobilized column, which required only one Galβ1,4-residue on glycopeptides for specific binding (Supplementary Fig. S1). The high sensitivity of the LTQ/Orbitrap Velos mass spectrometer could also compensate for the decreased amount of glycopeptides carrying Galβ1,4-terminated glycans in the sample mixture. Given these circumstances, the proteins identified in the β4GalT-I−/− mice, as well as those in the WT mice, were considered as true positives.
Our results indicated that the 70 glycoproteins identified solely from the WT mice were the plausible candidates for the β4GalT-I-specific target proteins. These proteins were indeed classified properly (based on the presence of Galβ1,4-terminated glycans on these proteins) owing to the careful means for purifying glycopeptides carrying Galβ1,4-terminated glycans from the WT and β4GalT-I−/− mice and then the stringent selection conditions to identify the candidate target proteins. Consistent with this notion, we found that these glycoproteins possess distinctive molecular features (Fig. 3,4). If these WT-only proteins were constituted of proteins that we failed to identify in the β4GalT-I−/− mice due to some technical or procedural limitations, then their molecular features should have been similar to those of the sharable targets. However, the molecular features of the glycoproteins identified solely from the WT mice were clearly distinct from those of the sharable target proteins. Therefore, we consider these proteins to be specific target proteins of β4GalT-I with high reliability, while further examinations are required to warrant the results.
There were 72 KO-only proteins whose molecular features were similar to those of the sharable target proteins. Judging from the similarity, we speculate that most of these KO-only proteins, if not all, were constituted of proteins unidentified in the WT mice. These KO-only proteins were not considered to be a result of chance detection, as these were detected in at least 3 (out of 12) independent LC/MS analyses of the β4GalT-I−/− samples and not detected even once in any WT samples. A potential reason for finding these proteins solely in the β4GalT-I−/− mice is diverse glycan alterations, rather than simple loss of Galβ1,4-residue from the glycan moiety of these proteins. One such possible alteration in glycan would be modifications of the terminal LacNAc motif. RCA120 is known to possess much reduced affinity for fucosylated glycans24. Thus, glycopeptides carrying a fucosylated glycan, such as Fucα1,2Galβ1,4GlcNAc- or Galβ1,4(Fucα1,3)GlcNAc-, would not bind to the RCA120-immobilized column even if they were present in the WT samples. The same core-peptides, however, can be identified in the β4GalT-I−/− samples because of decreased fucosylation. The decreased fucosylation in the β4GalT-I−/− mice could be explained as follows: some of the β4GalT isozymes, except β4GalT-I, are known to exhibit restricted branch-specificities for N-glycans in vitro16. In addition, β4GalT-I−/− mice express reduced β4GalT activity14,15. Thus, it is likely that the Galβ1,4-residue(s) would attach to a particular branch of N-glycans in β4GalT-I−/− mice. If a fucosyltransferase did not show any preference for the branches that are β1,4-galactosylated by the β4GalT isozyme, then the fucosylation of the LacNAc motif would decrease in β4GalT-I−/− mice. A differential identification pattern is also expected for the glycopeptides that carry α1,3-linked Gal-residue attached to the terminal Galβ1,4-residue(s). Furthermore, such alterations would not occur uniformly on all target proteins. Thus, we believe that the KO-only proteins were also β1,4-galactosylated in WT mice, but the RCA120-immobilized column could not capture them because of certain glycan modifications. Alternatively, the KO-only proteins were not identified in the WT mice because of their low abundance in the liver. However, these low-abundance proteins could be identified in the β4GalT-I−/− mice as a result of loss of or significant decrease in β1,4-galactosylation of high-abundance proteins. This is because the high-abundance proteins in the WT samples could significantly interfere with the detection of low-abundance proteins by limiting the detection dynamic-range during the LC/MS analysis. While both of these scenarios are possible explanation for the identification results, further detailed studies are required to reveal the underlying reason.
It is worth noting that the KO-only proteins also provided an important clue for unraveling the underlying mechanism of the phenotypes observed in the β4GalT-I−/− mouse. Altered glycan structures, including alterations other than the loss of Galβ1,4-residue, can disrupt the function of a protein carrying this altered glycan. Thus, identification of proteins found only in the β4GalT-I−/− mice is also informative. It would be essential to determine the structures of glycan moieties on the individual proteins to unravel the underlying mechanism for the phenotype as well as the rationale for the KO-only proteins. However, at present, it seems quite challenging to reveal the structure of the glycan in every individual protein on a proteome-scale due to lack of suitable technology.
Our approach is diversely applicable to other glycosyltransferases. For example, the target proteins specific for β1,3-N-acetylglucosaminyltransferase 2 (β3GnT2), which synthesizes poly-LacNAc in vivo8, could be identified in a manner similar to that described here. Peptides carrying poly-LacNAc could be specifically isolated from the WT and β3GnT2−/− mice using the Lycopersicon esculentum lectin-immobilized column32. Likewise, target proteins of a particular isozyme of sialyltransferases and fucosyltransferases could be identified by utilizing a plant lectin specific for sialylated glycans and fucosylated glycans, such as Maackia amurensis leukoagglutinin33 and Aleuria aurantia lectin34, respectively. These glycan motifs are also reported to play important roles in vivo35,36,37,38,39. The proteome-scale identification of the target proteins will provide a novel viewpoint to understand the roles of the glycans.
While β4GalT-I also catalyzes β1,4-galactosylation of O-glycans40, identification of β4GalT-I-specific target proteins carrying O-glycans still remains to be achieved. Our proteome-scale approach is inapplicable to identify the target proteins with O-glycans, because PNGase-F is used to incorporate 18O as a specific tag at the N-glycosylation site. However, our approach will facilitate unraveling the role of N-glycans through identification of target proteins of a wide variety of glycosyltransferase families and will also contribute to understanding of the complex mechanism that controls assembly of a particular glycan motif on specific proteins. The proteome-scale identification of target proteins of a glycosyltransferase isozyme provides a novel framework for future studies in the glycobiology field.
Methods
Sample preparation
The animal experiments were conducted according to the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan and approved by the Committee on Animal Experimentation of Kanazawa University. β4GalT-I−/− mice were created as described previously15. Livers (ca.1.0 g, wet weight) isolated from female mice (two from 30- and 31-week old WT mice; two from 32-week old β4GalT-I−/− mice) were separately cut into small pieces and then homogenized on ice. After sonication on ice, each homogenate was centrifuged at 29,000 g for 30 min at 4°C and the resulting supernatant was collected. Protein concentration in each supernatant was determined using the DC Protein assay kit (Bio-Rad) and bovine serum albumin as the standard protein. After bubbling N2 gas for 10 min, proteins in the supernatant were reduced by adding dithiothretiol (DTT) to a final DTT:protein ratio of 1:1 (wt:wt) and the solution was incubated at room temperature for 2 h under constant N2 gas bubbling condition. Proteins were then S-carbamoylmethylated with iodoacetamide (idoacetamide:protein = 2.5:1, wt:wt) in the dark for 2 h at room temperature under N2 gas atmosphere. The resulting solution was dialyzed against 10 mM Tris/HCl (pH 7.5) at 4°C, changing the dialysis buffer four times. Next, an aliquot of S-carbamoylmethylated proteins was digested overnight at 37°C with N-tosylphenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Thermo Scientific) using the enzyme:substrate ratios of 1:50 (wt:wt). Phenylmethylsulfonyl fluoride was added to a final concentration of 1 mM to terminate the digestion. Samples (before and after the protease reaction) were analyzed by SDS-PAGE (12%) and CBB-staining to ensure complete protease digestion.
Isolation of glycopeptides carrying Galβ1,4-terminated glycan using RCA120-lectin affinity chromatography
Tryptic digested liver extract (108 mg) was loaded onto an RCA120-immobilized column (LA-RCA120, 4.6 mm inner diameter (i.d.) × 150 mm, 16.6 mg of lectin/ml, J-Oil Mills) equilibrated with 10 mM Tris/HCl (pH 7.5), at a flow rate of 0.3 ml min−1. Before loading, we treated the tryptic digested liver extract with 25 mM HCl at 80°C for 1 h for desialylation. The desialylated glycopeptide mixture was neutralized with NaOH. Next, the column was washed with the binding buffer until the absorbance of the effluent at 230 nm was below 0.005 and then the glycopeptides were eluted off the column with the same buffer containing 50 mM lactose. To achieve maximal recovery of glycopeptides carrying Galβ1,4-terminated glycan, we reloaded the recovered flow-though fraction more than twice onto the same column and then the RCA120-bound glycopeptides were collected as described above. All glycoprotein containing fractions were combined and the glycopeptides were further purified by HILIC using a TSKgel Amide-80 column (2.0 mm i.d. × 50 mm, Tosoh). Briefly, acetonitrile (MeCN) and trifluoroacetic acid (TFA) were added to the pooled glycopeptides to adjust the final concentrations of MeCN and TFA to 70% (vol/vol) and 0.1% (vol/vol), respectively. This mixture was then loaded onto the TSKgel Amide-80 column, which was equilibrated with 70% MeCN and 0.1% TFA, at a flow rate of 0.1 ml min−1. After washing, the glycopeptides were eluted off the column with 50% MeCN in H2O containing 0.1% TFA and the absorbance of the effluent was monitored at 280 nm.
PNGase-F-mediated 18O-labeling of glycopeptides
N-glycosylated peptides were labeled specifically with 18O using the IGOT reaction as described previously25. Briefly, the HILIC-purified glycopeptides were concentrated in vacuo using a centrifugal concentrator. Then, 2 µl of 1 M Tris/HCl (pH 8.6) was added to the sample and it was dried again in vacuo using the centrifugal concentrator to completely remove the remaining H216O containing solvent. The dried glycopeptides were dissolved in H218O (98% atom% 18O, Taiyo Nippon Sanso Corp.), 2.3 mU of PNGase-F (Takara-Bio), dissolved in H218O, was then added to the solution and the mixture was subsequently incubated overnight at 37°C.
Nano-LC-MS/MS analysis of 18O-labeled peptides
The deglycosylated 18O-labeled peptide mixture was analyzed by an automated nanoflow HPLC coupled on-line via a nanoelectrospray ion source to an LTQ/Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Briefly, the peptide mixture was acidified with 1% formic acid and then injected into the reverse phase trap column (0.2 mm i.d. × 10 mm, MonoCap-C18 for trap, GL Science) of the LC system at a flow rate of 15 µl min−1. After washing with 0.1% formic acid for 15 min, the trap column was connected to the nano-flow LC system via the switching valve and the peptides were separated sequentially on a reverse phase tip column (150 µm i.d. × 70 mm, Mightysil-C18, 3 µm particles, Kanto Chemical) by applying a 0–35% linear gradient of MeCN in 0.1% (vol/vol) formic acid for 105 min at a flow rate of 100 nl min−1. The eluted peptides were electrosprayed directly into the LTQ/Orbitrap Velos mass spectrometer. Full MS scans were acquired in the Orbitrap with a resolution of 30,000 at m/z 400. The mass spectrometer was operated in the positive ion mode employing a data-dependent Top-10 method. In each Orbitrap survey scan, a full-scan spectrum was acquired in the Orbitrap in the mass range of m/z 450–1,500, followed by ion-trap CID on 10 most intense ions. The ion-trap MS analysis was performed with normalized collision energy for CID set at 35%, an activation q = 0.25 and an activation time of 10 ms for one microscan. The following conditions were used: spray voltage: 2.0 kV, ion transfer tube temperature: 200/250°C, ion selection threshold: 10,000 counts, maximum ion accumulation times: 500 ms for full scans, dynamic exclusion duration: 60 s (10 ppm window, maximum number of excluded peaks: 500).
Protein identification by database search
Raw data files were converted to MGF files using version 1.1 of the Protein Discoverer software (Thermo Fisher Scientific) and then processed using the version 2.2 of Mascot algorithm (Matrix Science) to match peptides by searching the NCBI mouse RefSeq protein sequence database (30,041 entries, downloaded on 25 Feb., 2011). The database-search was performed using the parameters described as follows. For the IGOT peptide identification, we included a custom modification, “deamidation with 18O (asparagine +2.98826 Da),” for the deamidation of 18O-incorporated asparagines. The sole fixed modification parameter was carbamoylmethylation (Cys) and the variable modification parameters were pyro-Glu (Gln at peptide N-terminus), oxidation (Met), deamination (carbamoylmethylated Cys at peptide N-terminus) and deamidation with 18O (Asn). Enzyme was set as trypsin with a maximum number of missed cleavages of 2 and a peptide molecular weight tolerance of 7 ppm. Peptide charges from +2 to +4 states and MS/MS tolerances of 0.8 Da were allowed. The significant threshold was set to 0.01 and the false discovery rate for each analysis was calculated by the Mascot algorithm using a decoy database. Peptide search results were exported as a CSV file and processed by Microsoft Excel. At first, we selected the peptides with rank 1 and expectation value within 0.01 as “identified peptides”. If a prospective “identified peptide” contained one or more 18O-labeled aspartic acid residue in consensus tripeptide sequence for N-glycosylation (Asn-Xaa-[Ser/Thr/Cys], where Xaa is any amino acid except proline), the peptide was accepted as an “N-glycopeptide”. We accepted Asn-Xaa-Cys as the consensus N-glycosylation sequence, as suggested by the previous report22. When the prospective “identified peptide” failed to meet these criteria, we considered the result as ambiguous and eliminated the data regardless of the match score. Since many glycoproteins possess only one N-glycosylation site, we also accepted a single peptide match result as an “identified glycoprotein” when we found in our analysis that the consensus N-glycosylation sequence Asn-Xaa-[Ser/Thr/Cys] was converted into Asp-Xaa-[Ser/Thr/Cys] with 18O-incorporation, especially in the light of high mass accuracy.
Prediction of membrane topology and classification of identified proteins
The presence of signal peptide in a protein was predicted using the web-based bioinformatics tool, SignalP 3.0 (Neural Network D-score and Hidden Markov models: http://www.cbs.dtu.dk/services/SignalP/)29. The predicted result was accepted only when both algorithms predicted identical results; otherwise, we considered the predicted result as “ambiguous”. We used version 2.0 of TMHMM (http://www.cbs.dtu.dk/services/TMHMM/)30 and version 1.1.0 of SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/sosuiframe0.html)31 for predicting TM segments. In this study, proteins predicted to contain a signal peptide and no TM segment were assigned as “soluble-type” and those predicted to contain TM segment(s) were assigned as “membrane-type” regardless of the outcome of the signal peptide prediction results. A possible signal peptide domain, which was predicted by TMHMM and SOSUI algorithms, was not counted as a TM domain when predicting protein localization.
References
Varki, A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 (1993).
Ohtsubo, K. & Marth, J. Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 (2006).
Varki, A. et al. Essential of Glycobiology. 2nd edition. (Cold Spring Harbor Laboratory Press, New York, 2009).
Lowe, J. & Marth, J. A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 72, 643–691 (2003).
Sarkar, M. et al. Null mutations in Drosophila N-acetylglucosaminyltransferase I produce defects in locomotion and a reduced life span. J. Biol. Chem. 281, 12776–12785 (2006).
Schachter, H. Protein glycosylation lessons from Caenorhabditis elegans. Curr. Opin. Struct. Biol. 14, 607–616 (2004).
Taniguchi, N. et al. Decoding sugar functions by identifying target glycoproteins. Curr. Opin. Struct. Biol. 16, 561–566 (2006).
Togayachi, A. et al. Polylactosamine on glycoproteins influences basal levels of lymphocyte and macrophage activation. Proc. Natl. Acad. Sci. USA 104, 15829–15834 (2007).
Demetriou, M., Granovsky, M., Quaggin, S. & Dennis, J. W. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409, 733–773 (2001).
Shaper, N. et al. Bovine galactosyltransferase: identification of a clone by direct immunological screening of a cDNA expression library. Proc. Natl. Acad. Sci. USA 83, 1573–1577 (1986).
Narimatsu, H., Sinha, S., Brew, K., Okayama, H. & Qasba, P. Cloning and sequencing of cDNA of bovine N-acetylglucosamine (beta 1-4)galactosyltransferase. Proc. Natl. Acad. Sci. USA 83, 4720–4724 (1986).
Taniguchi, N., Honke, K. & Fukuda, M. (Eds.) Handbook of Glycosyltransferases and Related Genes. (Springer, Tokyo, 2002).
Nairn, A. V. et al. Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J. Biol. Chem. 283, 17298–17313 (2008).
Lu, Q., Hasty, P. & Shur, B. Targeted mutation in beta1,4-galactosyltransferase leads to pituitary insufficiency and neonatal lethality. Dev. Biol. 181, 257–267 (1997).
Asano, M. et al. Growth retardation and early death of beta-1,4-galactosyltransferase knockout mice with augmented proliferation and abnormal differentiation of epithelial cells. EMBO J. 16, 1850–1857 (1997).
Ito, H. et al. Strategy for the fine characterization of glycosyltransferase specificity using isotopomer assembly. Nat. Methods 4, 577–582 (2007).
Asano, M. et al. Impaired selectin-ligand biosynthesis and reduced inflammatory responses in beta-1,4-galactosyltransferase-I-deficient mice. Blood 102, 1678–1685 (2003).
Mori, R., Kondo, T., Nishie, T., Ohshima, T. & Asano, M. Impairment of skin wound healing in beta-1,4-galactosyltransferase-deficient mice with reduced leukocyte recruitment. Am. J. Pathol. 164, 1303–1314 (2004).
Nishie, T. et al. Development of immunoglobulin A nephropathy-like disease in beta-1,4-galactosyltransferase-I-deficient mice. Am. J. Pathol. 170, 447–456 (2007).
Steentoft, C. et al. Mining the O-glycoproteome using Zinc-finger nuclease glycoengineered SimpleCells. Nat. Methods 8, 977–982 (2011).
Schjoldager, K. T. et al. Probing isoform-specific functions of polypeptide GalNAc-transferase using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. USA 109, 9893–9898 (2012)
Zielinska, D., Gnad, F., Wiśniewski, J. & Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 (2010).
Green, E. D., Brodbeck, R. M. & Baenziger, J. U. Lectin affinity high-performance liquid chromatography. Interactions of N-glycanase-released oligosaccharides with Ricinus communis agglutinin II. J. Biol. Chem. 262, 12030–12039 (1987).
Itakura, Y. et al. Systematic comparison of oligosaccharide specificity of Ricinus communis agglutinin I and Erythrina lectins: a search by frontal affinity chromatography. J. Biochem. 142, 459–469 (2007).
Kaji, H. et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 21, 667–672 (2003).
Kaji, H., Yamauchi, Y., Takahashi, N. & Isobe, T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat. Protoc. 1, 3019–3027 (2006).
Kaji, H. et al. Proteomics reveals N-linked glycoprotein diversity in Caenorhabditis elegans and suggests an atypical translocation mechanism for integral membrane proteins. Mol. Cell. Proteomics 6, 2100–2109 (2007).
An, Y. & Cipollo, J. F. An unbiased approach for analysis of protein glycosylation and application to influenza vaccine hemagglutinin. Anal. Biochem. 415, 67–80 (2011).
Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Hirokawa, T., Boon-Chieng, S. & Mitaku, S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 (1998).
Nachbar, M. & Oppenheim, J. Tomato (Lycopersicon esculentum) lectin. Methods Enzymol. 83, 363–368 (1982).
Wang, W. C. & Cummings, R. D. The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked alpha-2,3 to penultimate galactose residues. J. Biol. Chem. 263, 4576–4585 (1988).
Yazawa, S., Furukawa, K. & Kochibe, N. Isolation of fucosyl glycoproteins from human erythrocyte membranes by affinity chromatography using Aleuria aurantia lectin. J. Biochem. 96, 1737–1742 (1984).
Wang, X., Gu, J., Miyoshi, E., Honke, K. & Taniguchi, N. Phenotype changes of Fut8 knockout mouse: core fucosylation is crucial for the function of growth factor receptor(s). Methods Enzymol. 417, 11–22 (2006).
Kudo, T. et al. Mice lacking alpha1,3-fucosyltransferase IX demonstrate disappearance of Lewis x structure in brain and increased anxiety-like behaviors. Glycobiology 17, 1–9 (2007).
Malý, P. et al. The alpha(1,3)fucosyltransferase Fuc-TVII controls leukocyte trafficking through an essential role in L-, E- and P-selectin ligand biosynthesis. Cell 86, 855–867 (2006).
Hedlund, M., Ng, E., Varki, A. & Varki, N. M. alpha 2-6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 68, 388–394 (2008).
Hennet, T., Chui, D., Paulson, J. C. & Marth, J. D. Immune regulation by the ST6Gal sialyltransferase. Proc. Natl. Acad. Sci. USA 95, 4504–4509 (1998).
Kotani, N., Asano, M., Iwakura, Y. & Takasaki, S. Impaired galactosylation of core 2 O-glycans in erythrocytes of beta1,4-galactosyltransferase knockout mice. Biochem. Biophys. Res. Commun. 260, 94–98 (1999).
Acknowledgements
We thank T. Ohkura, T. Satoh and H. Sawaki of National Institute of Advanced Industrial Science and Technology for fruitful discussions. We also thank A. Takahashi for assistance in the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
D.S. conceived and designed the study, performed most of the experiments, analyzed data and wrote the paper with comments from H.K. and H.N.; H.K. conceived and designed the study, performed LC/MS analyses; K.S. and M.A. developed β4GalT-I−/− mice and provided its sample tissues; H.N. conceived and designed the study and supervised all aspects of the work; and all authors discussed the results and implications and commented on the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary dataset
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Sugahara, D., Kaji, H., Sugihara, K. et al. Large-scale identification of target proteins of a glycosyltransferase isozyme by Lectin-IGOT-LC/MS, an LC/MS-based glycoproteomic approach. Sci Rep 2, 680 (2012). https://doi.org/10.1038/srep00680
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00680
This article is cited by
-
Identification of Poly-N-Acetyllactosamine-Carrying Glycoproteins from HL-60 Human Promyelocytic Leukemia Cells Using a Site-Specific Glycome Analysis Method, Glyco-RIDGE
Journal of the American Society for Mass Spectrometry (2018)
-
A peptide N-terminal protection strategy for comprehensive glycoproteome analysis using hydrazide chemistry based method
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.