Abstract
The discovery of new chlorophyllous pigments would provide greater understanding of the mechanisms and evolution of photosynthesis. Bacteriochlorophyll f has never been observed in nature, although this name was proposed ~40 years ago based on structurally related compounds. We constructed a bacteriochlorophyll f–accumulating mutant of the green sulfur bacterium Chlorobaculum limnaeum, which originally produced bacteriochlorophyll e, by knocking out the bchU gene encoding C-20 methyltransferase based on natural transformation. This novel pigment self-aggregates in an in vivo light-harvesting antenna, the chlorosome and exhibits a Qy peak of 705 nm, more blue-shifted than any other chlorosome reported so far; the peak overlaps the maximum (~700 nm) of the solar photon flux spectrum. Bacteriochlorophyll f chlorosomes can transfer light energy from core aggregated pigments to another bacteriochlorophyll in the chlorosomal envelope across an energy gap of ~100 nm and is thus a promising material for development of new bioenergy applications.
Similar content being viewed by others
Introduction
Chlorophyllous pigments are required by photosynthetic organisms in order to carry out primary production: harvesting sunlight energy, driving photosystems and achieving charge separation. However, the specific pigments present in the cells of a photosynthetic organism influence its light-harvesting efficiency, because each pigment species has distinctive absorption characteristics. Most oxygenic phototrophs, such as plants and cyanobacteria, contain chlorophyll (Chl) a, whereas some plants have Chl b, and some cyanobacteria possess Chl d1 and Chl f2. Differences among these Chls include oxidation of peripheral substituents of the chlorin ring structure. Chls b (Qy and Soret maxima = 642.2 and 453.0 nm in diethyl ether), d (Qy/Soret = 685.8/445.8 nm) and f (Qy/Soret = 694.5/439.5 nm)3 are differentiated from Chl a (Qy/Soret = 660.8/429.6 nm) by the presence of a formyl group at the C-7, C-3 and C-2 positions, respectively. Changes in the peripheral substituents of the pigment ring alter visible light absorbance; the different Chls confer habitat- or niche-specific selective advantages on photosynthetic organisms that possess them.
Green sulfur bacteria (GSBs) are strict anaerobes that utilize reduced sulfur compounds (sulfide, polysulfide, or elemental sulfur) as electron donors in anoxygenic photosynthesis. They thrive in deep layers of the photic zone, where reduced sulfur compounds are abundant while light is limited. GSBs contain characteristic light-harvesting organelles, called chlorosomes, that contain large quantities (1.4–2.1×105 molecules per chlorosome) of bacteriochlorophylls (BChls) c, d, or e (see Fig. 1C)4,5,6. The Qy and Soret absorption bands of BChls c (Qy/Soret = 660.4/432.0 nm in diethyl ether), d (Qy/Soret = 649.6/425.4 nm) and e (Qy/Soret = 646.4/459.6 nm) in their monomeric states overlap those of Chls a/b/d/f in oxygenic phototrophs living in upper layers of the photic zone. However, BChls c/d/e can form self-aggregated structures in chlorosomes7,8, resulting in red-shifted absorption bands: typical chlorosomal Qy/redmost Soret absorption maxima are 750–760/460 nm for BChl c, 730/450 for BChl d and 710–720/520 for BChl e (Fig. 2a). GSBs, therefore, can utilize light energy at greater depths than other organisms possessing Chls. Fluorescence of free Chl a in a solution is completely quenched at 0.3 M, a phenomenon called concentration quenching9. Although the concentration of BChls in chlorosome is much larger [~2 M, based on data in ref 4], no concentration quenching is observed. Moreover, chlorosomal BChl concentrations are also larger than those in other photosynthetic pigment-protein antenna systems, e.g., 0.3 M for photosystem II light-harvesting complex of higher plants10 and 0.1 M for Fenna-Matthews-Olson (FMO) BChl a protein in GSB11. These observations suggest that the well-ordered supramolecular structures of chlorosomal self-aggregated pigments permit rapid and efficient migration of excitation energy within chlorosomes, allowing GSB to efficiently absorb light in ecological niches with illumination conditions as low as 3–10 nE·s−1·m−212. Elucidation of the structural and photochemical mechanisms of chlorosomes would therefore be useful in developing bioenergy applications.
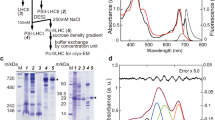
Determination of compositions of BChl f homologs in C. limnaeum bchU mutant.
(a) HPLC elution profiles of extracted pigments from the C. limnaeum wildtype (upper panel) and bchU mutant cells (bottom panel). The elution of pigments was detected at 455 nm. Peak 1, R[E,E]BChl e; peak 2, S[E,E]BChl e; peak 3, R[P,E]BChl e; peak 4, S[P,E]BChl e; peak 5, S[I,E]BChl e; Peak 6, R[E,E]BChl f; peak 7, R[P,E]BChl f; peak 8, S[P,E]BChl f; peak 9, S[I,E]BChl f. (b) UV-Vis absorption (upper) and CD (bottom) spectra of R[E,E]BChl f in diethyl ether. (c) Chemical structure of BChl f, with comparison to other chlorosomal BChls.
The name BChl f was proposed ~40 years ago to denote the 20-unsubstituted form of BChl e (Fig. 1c)13. The structural relationship between BChl e and f is the same as that between BChl c and d, namely, a difference of a methyl or hydrogen group at C-20 position. Methylation at C-20 of BChl c is catalyzed by the gene product of the bchU gene14,15. The GSB Chlorobaculum (C.) parvum NCIB 8327 has a natural loss-of-function mutation in bchU14,16; therefore, this strain produces only BChl d, but it can be altered by a second mutation to produce BChl c under low-light conditions17,18. Restored production of BChl c in the strain under low light illumination was ascribable to that self-aggregated BChl d in chlorosomes was less efficient in energy transfer to the baseplate proteins in chlorosomal envelope19. Because such spontaneous mutation in bchU occurs frequently, we hypothesized that a GSB that naturally produces BChl e could be easily altered to produce BChl f. However, although BChl d is common20,21, BChl f has never been detected in nature. A BChl f homolog was synthesized by modifying Chl b; it is stable in vitro, and its chemical and absorption properties have been characterized22. Using synthetic BChl f as a standard, detailed HPLC analyses were conducted on GSBs that produce BChl e; however, no natural BChl f molecule has previously been found. The absence of natural BChl f suggested that such a mutation in a BChl e–producing strain would dramatically reduce light harvesting and energy transfer, conferring a selective disadvantage on mutants producing BChl f. This prediction has heretofore not been tested, largely because BChl e–producing GSBs are difficult to culture and genetically manipulate.
We have serially cultured the BChl e–producing GSB strain C. limnaeum 1549 for about a decade and have isolated a derivative strain that grows on plates (hereafter, called ‘RK-j-1’). We performed insertional inactivation of bchU in RK-j-1 strain, yielding a mutant that produces BChl f. Here, we report the absorption properties of the resulting mutant and characterize its chlorosomes, which are composed of self-aggregated BChl f. This work was initially submitted as a patent on February 13, 201223. While the present paper was under review, Bryant's group reported similar results independently24 (see also Supplementary information).
Results
Identification of BChl f homologs in C. limnaeum bchU mutant
Mutation of bchU was achieved based on natural transformation and homologous recombination in the similar way previously described for the BChl c–producing bacterium C. tepidum25 (Fig. S1). Pigments extracted from wildtype and mutant RK-j-1 cells were analyzed by HPLC (Fig. 1a). Elution profiles of pigment extracts from wildtype contained five BChl e homologs possessing an R or S stereo-configuration at the C-31 asymmetric position and different degrees of methylation at C-82 (Figures 1a upper and 1c): (31R)-epimeric 8,12-diethyl-(R[E,E])BChl e (peak 1), (31S)-epimeric 8,12-diethyl-(S[E,E])BChl e (peak 2), (31R)-epimeric 8-propyl-12-ethyl-(R[P,E])BChl e (peak 3), (31S)-epimeric 8-propyl-12-ethyl-(S[P,E])BChl e (peak 4) and (31S)-epimeric 8-isobutyl-12-ethyl-(S[I,E])BChl e (peak 5). The assignments for the R or S configuration at the C-31 position were based on the elution pattern described previously22. In HPLC profiles of mutant cells, four peaks were detected (Fig. 1a, lower), with the same online electronic absorption spectra: λmax = 639 and 456 nm in 70:15:15 (v/v/v) acetonitrile:acetone:water. These spectra are consistent with that of the authentic BChl f standard22. The four bands were 14 Da less massive than the corresponding peaks in wildtype cells (Table S2). These observations established that peaks 6, 7, 8 and 9 were assigned as R[E,E]-, R[P,E]-, S[P,E]- and S[I,E]BChl f, respectively. The UV-Vis absorption spectrum of isolated R[E,E]BChl f (the main peak 6) in diethyl ether exhibits a 12.6-nm blue-shifted Qy maximum at 633.2 nm and an 8.8-nm blue-shifted Soret maximum at 449.6 nm, in contrast to those of R[E, E]BChl e (Fig. 1b upper), which are identical to those of the R/S[E, M]BChl f standard22. The circular dichroism (CD) spectrum of R[E,E]BChl f exhibits blue-shifted peak positions relative to R[E,E]BChl e (Fig. 1b lower); intensities of the e peaks were higher than those of f. Furthermore, we determined the molecular structure of R[E,E]BChl f using 1H nuclear magnetic resonance (NMR) spectroscopy (Fig. S2). Based on these data, we conclude that the bchU-inactivated mutant exclusively accumulates BChl f homologs and BChl e species are not present in mutant cells.
Absorption properties of BChl f self-aggregation in vivo
In the UV-Vis-NIR absorption spectrum of bchU mutant cells, we observed the Qy peak at 704.8 nm and the Soret band at ~510 nm (Fig. 2a). These in vivo absorption bands were red-shifted by ~70 and 60 nm from those of monomeric BChl f (Fig. 1b upper), indicating that BChl f homologs formed self-aggregates in chlorosomes. The Qy and redmost Soret absorption band in the mutant cells were 15-nm blue-shifted compared with wildtype, but the Soret bands on the blue side were almost identical (λmax = 450.2 nm) (Fig. 2a). These spectroscopic relationships between BChl e and f were very similar to those between BChl c and d26. Notably, the BChl f-aggregates in chlorosomes exhibited the most blue-shifted Qy peak reported so far among chlorosomes in vivo. Such self-aggregates in a BChl f-chlorosome have supramolecular structures similar to those in the corresponding BChl e-chlorosome, as revealed by similarities between the shapes of the CD spectra of bchU mutant and wildtype cells (Fig. 2b); however, the mutant peak positions were blue-shifted relative to the wildtype, except for the 440-nm positive peak.
Photosynthetic properties of BChl f-chlorosomes
The NIR bands at ~804 nm, observed in both wild and bchU mutant cells, were primarily derived from BChl a molecules in the chlorosomal baseplate and from FMO proteins as the first and second acceptors of excitation energy from chlorosomal core BChl aggregates, respectively (Fig. 2a). In the corresponding isolated chlorosomes lacking FMO proteins, 10-nm–blue-shifted bands at 793.0 nm (Fig. S3) were ascribed to BChl a molecules in baseplate proteins, while a small shift was observed in major chlorosomal Qy peaks (704.8 to 703.0 nm). Thus, we estimated the energy gap between BChl f-aggregates and BChl a to be 90–100 nm (1600–1800 cm−1), larger than that in wildtype chlorosomes. To clarify whether chlorosomal self-aggregates of BChl f in the bchU mutant are photosynthetically active over such a large energy gap, we measured fluorescence emission spectrum of the mutant cells (Fig. 2a inset). We observed singlet excited energy transfer from BChl f-aggregates (λem = 723 nm) to the 810-nm emission species in mutant chlorosomes, similar to the transfer from BChl e to BChl a in the wildtype. The 810-nm emission species is ascribed to BChl a in baseplate and/or FMO proteins and therefore, we could not discuss here about efficiency of net energy transfer from BChl f-aggregates to baseplate-BChl a which is next energy acceptor. Although the energy gap in the mutant was larger than in the wildtype, the energy transfer did indeed occur. Growth measurements of the bchU mutant cells at several light intensities supported this result; the more intense the illumination was, the faster the mutant grew (Table 1 and Fig. S4). Thus, we conclude that the bchU mutant could grow via phototrophic activity of BChl f, although the mutant grew 20–30% more slowly than wildtype under all conditions examined. The suppression of mutant growth rate could be due to Förster-type energy transfer in the chlorosomes19.
Discussion
Here we reported that the first time BChl f has been observed in vivo. If the bchU mutant can grow using photosynthetically active BChl f molecules, why has this pigment been never found in nature? Ecological studies revealed that BChl e–containing species predominate at the lowest depth in the photic zone, e.g., at 100 m in the Black Sea12. In such a region, almost all available light is green-yellow (~530 nm)21; BChl e–containing species, which possess a Soret band at ~520 nm, can utilize this light energy more efficiently than BChl c– or d–containing species. In contrast, BChl f-chlorosomes, with a Soret band at ~510 nm, capture this light less efficiently. Beyond this disadvantage in light-harvesting efficiency, self-aggregates of BChl f exhibit less effective energy transfer to BChl a due to their large energy gap. Therefore, BChl f–containing bacteria might find it very difficult to survive, especially in habitats where BChl e–producing bacteria are already predominant.
As in the study of C. parvum and C. tepidum14,16 that demonstrated BchU's role in converting BChl d to BChl c, we have demonstrated that in C. limnaeum BchU catalyzes conversion of BChl e from BChl f species. Among the chlorosomal pigments (BChls c/d/e), the biosynthetic pathway of BChl e is not well understood, especially regarding the formylation at C-7 (oxidation of a methyl group to formyl at C-7). Since the plating strain of C. limnaeum, RK-j-1, is now amenable to genetic manipulation, it will be possible to test genes potentially encoding enzyme(s) involved in this oxidation. After the pathway of BChl e has been characterized, it will be useful to create isogenic mutants that produce each of the BChls c/d/e/f individually or in different combinations. Because of the inherent compositional and morphological heterogeneity of chlorosomes, the structural arrangements of chlorosomal self-aggregates have not yet been determined; several distinct structural models have been proposed27,28,29. Therefore, structural studies on chlorosomes using mutants producing BChl c, d, e, and/or f will facilitate understanding of the bioenergetics of GSBs. Furthermore, elucidating the energetics of BChl f-chlorosomes, which exhibit the largest energy gap among all known phototroph chlorosomes, will be useful in developing artificial photosynthetic devices, capable of being driven by a wide range of sunlight energies.
Methods
Bacterial strains and culture conditions
We continuously subcultured Chlorobaculum limnaeum 154922 for about ten years and obtained a derivative strain, RK-j-1, used as wildtype in this study. This bacterium was anaerobically grown in screw-capped bottles filled with liquid CL media, or on solid CP plates6.
For molecular techniques, E. coli DH5α was grown in LB medium containing 100 μg/mL of ampicillin or 10 μg/mL gentamycin, as appropriate.
Determination of DNA sequences of the flunking region of bchU gene in C. limnaeum
A partial DNA sequence of the bchU gene from C. limnaeum 1549 was determined by Maresca et al.14 (Accession No. AY452767) and used for designing primers for inverse PCR amplified genes containing unknown regions of bchU and its flanking genes. The genomic DNA of C. limnaeum was extracted and digested by restriction enzyme HindIII. The resulting DNA fragments were self-ligated and used as the template for inverse PCR with a primer set of inverseU F and R (Table S1). An about 9-kbp DNA fragment containing bchU was amplified by the inverse PCR and then the complete bchU gene and its franking regions (1.9-kbp) were sequenced with an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, California, USA) (Accession No. AB721963).
Plasmid constructions
The PCR primers used in this study for the construction of plasmids are described in Table S1. To construct the C. limnaeum mutant lacking bchU, the plasmid pUCbchUGm (Fig. S1A) was produced as follows. A 1.53-kbp blunt-ended DNA fragment containing the bchU gene was amplified from the genomic DNA of C. limnaeum using KOD -plus- DNA polymerase (TOYOBO, Osaka, Japan) and primers Uaround F and Uaround R. The PCR product was digested at the EcoRI restriction enzyme site designed in Uaround R primer and cloned into the SmaI and EcoRI sites of pUC118 (TAKARA, Shiga, Japan), yielding pUCbchU plasmid. To disrupt the unique EcoRI and BamHI sites in pUCbchU for after cloning step, this plasmid was digested by EcoRI, blunted and self-ligated and the resulting plasmid was in turn treated with BamHI, blunted and self-ligated, producing pUCbchUEB. The aacC1 gene, conferring resistance to gentamycin, was excised from pUCGM30 using SacI. After blunting, the gene fragment was sub-cloned into the SmaI site of pUC118. From the resulting plasmid, pUCGMF, the aacC1 gene was excised by the treatment with BamHI and EcoRI. To amplify the DNA fragment from pUCbchUEB without the inner portion of bchU, the plasmid was used as the template for PCR with primers (the directions of primers shown in Fig. S1A). The resulting PCR product was digested by BamHI and EcoRI, (designed in the phaeU F and R primers, respectively) and ligated with the excised aacC1 gene fragment, yielding pUCTbchUGm.
Natural transformation of C. limnaeum RK-j-1
The standard protocol for natural transformation was essentially according to the method described for C. tepidum by Frigaard and Bryant7. Cells from 3 mL of full-growth culture of C. limnaeum RK-j-1 were harvested in a microcentrifuge tube and resuspended in 100 μL of fresh CL medium. The cell suspension was mixed with ≥ 0.1 mg of HindIII-digested pUCbchUGm and the mixture was spotted on a CP plate containing no antibiotics. To dry the spot, the plate was incubated in dark for 1 hour at room temperature. Next, the plate was placed upside-down in an anaerobic jar (#HP0031A, OXOID, Hamphire, UK) with catalysis (#BR0042A), an Anaerobic Gas Generating Kit (#BR0038B) and a small tube containing 100 mg of thioacetamide dissolved in 1 mL of aqueous 1 M HCl. The jar was kept in the dark at room temperature for 6–12 hours and then incubated under illumination at 30°C for 5–7 days. Cells in each spot were scraped off and suspended in 1 mL of CL medium; the cell suspension was diluted and spread on CP plates containing 50 μg/mL of gentamicin. The plates were placed in jar the same manner as described above and incubated for 2–3 weeks to allow single colonies to appear.
PCR analysis was carried out to monitor segregation of wildtype and mutant alleles using primers limnI comf. F and R. A DNA molecular weight marker, λ/StyI digest (TOYOBO, Osaka, Japan), was used for molecular mass estimations of PCR products. Furthermore, DNA sequence analysis of the PCR fragments was performed using primers Uaround F, Uaround R, seq1 and seq2.
Determination of compositions of BChl f homologs
Pigments of the C. limnaeum mutant strain were extracted and analyzed as follows. A mixture of acetone and methanol (9:1, v/v) was added to the harvested cells and mixed using a vibrator. A mixture of diethyl ether and petroleum ether (1:1, v/v) and then distilled water were added to transfer the pigment components to the ether layer. The ether phase was collected and evaporated to dryness under a stream of N2 gas and the residues were dissolved in a small amount of HPLC eluent, described below. HPLC analysis was performed using a Shimadzu LCMS-2010EV system (Shimadzu, Kyoto, Japan) comprising a liquid chromatograph (SCL-10Avp system controller, LC-10ADvp pump and SPD-M10Avp photodiode-array detector) and a quadrupole mass spectrometer equipped with an atmospheric pressure chemical ionization (APCI) probe. Reverse-phase HPLC was performed under the following conditions: column, Cosmosil 5C18-AR-II (6.0 φ x 250 mm, Nacalai Tesque, Kyoto, Japan); eluent, acetonitrile:acetone:H2O (70:15:15, v/v/v); flow rate, 1.25 mL/min; detection wavelength, 455 nm. APCI-MS spectra were measured as follows: resolution, ±0.15 Da; capillary temperature, 250°C; APCI vaporizer temperature, 400°C; ionization voltage, 4.5 kV; sheath gas flow, 2.5 L/min; drying gas pressure, 0.02 MPa.
Isolation and structural determination of R[E,E]BChl f
Pigment components were extracted from harvested C. limnaeum mutant cells as described above. R[E,E]BChl f, a major homolog of BChl f (corresponding to peak #6 in Fig. 1a lower), was purified by HPLC as follows: column, Cosmosil 5C18-AR-II (10 φ x 250 mm, Nacalai Tesque); eluent, methanol:H2O (95:5, v/v); flow rate, 1.5 mL/min. The 600 MHz 1H-NMR spectra of the isolated R[E,E]BChl f were recorded in tetrahydrofuran(THF)-d8 (Euriso-top, Saclay, France) using a JEOL ECA-600 NMR spectrometer (JEOL, Akishima, Japan); the residual proton at the 3-position of THF-d7 (δH = 1.72 ppm) was used as an internal standard. A set of assignments of 1H signals was obtained from correlation spectroscopy (COSY) and rotating frame Overhauser enhancement spectroscopy (ROESY) (τm = 400 msec).
Spectroscopy measurements of bacterial cells, isolated chlorosomes and pigments
UV-Vis-NIR absorption spectra were measured using a Hitachi U-3500 (Hitachi High-Tech, Tokyo, Japan). Steady-state fluorescence emission spectra were measured using a C9920-03G series fluorescence measurement system (Hamamatsu Photonics, Shizuoka, Japan). CD spectra were observed with a Jasco J-720W spectropolarimeter (Jasco, Tokyo, Japan). Cells of C. limnaeum strains grown in the early stationary phase under irradiation ~30 μE·s−1·m−2 were collected, suspended in 50 mM Tris-HCl (pH 8.0) and used for UV-Vis-NIR absorption and CD spectra measurements. For fluorescence spectrum measurements, the cell suspension was mixed with 20 mM sodium dithionite and incubated for 30 min. Isolation of chlorosomes from 1 L full growth culture of the C. limnaeum wildtype and bchU mutant proceeded as previously described31,32.
Growth rate measurements of C. limnaeum bchU mutant
Pre-cultures of the wildtype and bchU mutant of C. limnaeum were grown to stationary phase under irradiation at ~30 μE·s−1·m−2. Three hundred μL aliquots of pre-cultures were inoculated into 30 mL screw-capped test tubes filled with freshly prepared CL media and cultures were then grown under illumination of 3, 30, or 130 μE·s−1·m−2 at 30°C. Optical cell densities were measured at 660 nm using a Mini Photo 518R photometer (TAITEC, Saitama, Japan).
References
Miyashita, H. et al. Chlorophyll d as a major pigment. Nature 383, 402 (1996).
Chen, M. et al. A red-shifted chlorophyll. Science 329, 1318–1319 (2010).
Li, Y., Scales, N., Blankenship, R. E., Willows, R. D. & Chen, M. Extinction coefficient for red-shifted chlorophylls: Chlorophyll d and chlorophyll f. Biochim Biophys Acta 1817, 1292–1298 (2012).
Martinez-Planells, A. et al. Determination of the topography and biometry of chlorosomes by atomic force microscopy. Photosynth Res 71, 83–90 (2002).
Montano, G. A. et al. Characterization of Chlorobium tepidum chlorosomes: a calculation of bacteriochlorophyll c per chlorosome and oligomer modeling. Biophys J 85, 2560–2565 (2003).
Saga, Y., Shibata, Y., Itoh, S. & Tamiaki, H. Direct counting of submicrometer-sized photosynthetic apparatus dispersed in medium at cryogenic temperature by confocal laser fluorescence microscopy: estimation of the number of bacteriochlorophyll c in single light-harvesting antenna complexes chlorosomes of green photosynthetic bacteria. J Phys Chem C 111, 12605–12609 (2007).
Frigaard, N. U. & Bryant, D. A. Seeing green bacteria in a new light: genomics-enabled studies of the photosynthetic apparatus in green sulfur bacteria and filamentous anoxygenic phototrophic bacteria. Arch Microbiol 182, 265–276 (2004).
Saga, Y., Shibata, Y. & Tamiaki, H. Spectral properties of single light-harvesting complexes in bacterial photosynthesis. J Photochem Photobiol C 11, 15–24 (2010).
Beddard, G. S. & Porter, G. Concentration quenching in chlorophyll. Nature 260, 366 (1976).
Kuhlbrandt, W. & Wang, D. N. Three-dimensional structure of plant light-harvesting complex determined by electron crystallography. Nature 350, 130–134 (1991).
Matthews, B. W., Fenna, R. E., Bolognesi, M. C., Schmid, M. F. & Olson, J. M. Structure of a bacteriochlorophyll a-protein from the green photosynthetic bacterium Prosthecochloris aestuarii. J Mol Biol 131, 259–285 (1979).
Overmann, J., Cypionka, H. & Pfennig, N. An extremely low-light-adapted phototrophic sulfur bacterium from the Black Sea. Limnol Oceanogr 37, 150–155 (1992).
Gloe, A., Pfennig, N., Brockmann, H., Jr & Trowitzsch, W. A new bacteriochlorophyll from brown-colored Chlorobiaceae. Arch Microbiol 102, 103–109 (1975).
Maresca, J. A. et al. The bchU gene of Chlorobium tepidum encodes the c-20 methyltransferase in bacteriochlorophyll c biosynthesis. J Bacteriol 186, 2558–2566 (2004).
Harada, J., Saga, Y., Yaeda, Y., Oh-Oka, H. & Tamiaki, H. In vitro activity of C-20 methyltransferase, BchU, involved in bacteriochlorophyll c biosynthetic pathway in green sulfur bacteria. FEBS Lett 579, 1983–1987 (2005).
Harada, J., Saga, Y., Oh-oka, H. & Tamiaki, H. Different sensitivities to oxygen between two strains of the photosynthetic green sulfur bacterium Chlorobium vibrioforme NCIB 8327 with bacteriochlorophyll c and d. Photosynth Res 86, 137–143 (2005).
Saga, Y., Oh-oka, H., Hayashi, T. & Tamiaki, H. Presence of exclusively bacteriochlorophyll-c containing substrain in the culture of green sulfur photosynthetic bacterium Chlorobium vibrioforme strain NCIB 8327 producing bacteriochlorophyll-d. Anal Sci 19, 1575–1579 (2003).
Broch-Due, M. & Ormerod, J. G. Isolation of a Bchl c mutant from Chlorobium with Bchl d by cultivation at low light intensity. FEMS Microbiol Lett 3, 305–308 (1978).
Causgrove, T. P., Brune, D. C. & Blankenship, R. E. Förster energy transfer in chlorosomes of green photosynthetic bacteria. J Photochem Photobiol B 15, 171–179 (1992).
Borrego, C. M., Garcia-Gil, J., Cristina, X. P., Vila, X. & Abella, C. A. Occurrence of new bacteriochlorophyll d forms in natural population of green photosynthetic sulfur bacteria. FEMS Microbiol Ecol 26, 257–267 (1998).
Vila, X. & Abella, C. A. Light-harvesting adaptations of planktonic phototrophic micro-organisms to different light quality conditions. Hydrobiolagia 452, 15–30 (2001).
Tamiaki, H. et al. In vitro synthesis and characterization of bacteriochlorophyll-f and its absence in bacteriochlorophyll-e producing organisms. Photosynth Res 107, 133–138 (2011).
Harada, J., Noguchi, M. & Tamiaki, H. Mutant of photosynthetic green bacterium and bacteriochlorophyll. Japan Patent JP 2012–028919 (2012).
Vogl, K., Tank, M., Orf, G. S., Blankenship, R. E. & Bryant, D. A. Bacteriochlorophyll f: properties of chlorosomes containing the "forbidden chlorophyll". Front Microbiol 3, 10.3389/fmicb.2012.00298 (2012).
Frigaard, N. U. & Bryant, D. A. Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl Environ Microbiol 67, 2538–2544 (2001).
Saga, Y. & Tamiaki, H. Comparison between chlorosomes containing bacteriochlorophyll-c and chlorosomes containing bacteriochlorophyll-d isolated from two substrains of green sulfur photosynthetic bacterium Chlorobium vibrioforme NCIB 8327. J Photochem Photobiol B 75, 89–97 (2004).
Staehelin, L. A., Golecki, J. R. & Drews, G. Supramolecular organization of chlorosomes (chlorobium vesicles) and of their membrane attachment sites in Chlorobium limicola. Biochim Biophys Acta 589, 30–45 (1980).
Psencik, J. et al. Lamellar organization of pigments in chlorosomes, the light harvesting complexes of green photosynthetic bacteria. Biophys J 87, 1165–1172 (2004).
Ganapathy, S. et al. Alternating syn-anti bacteriochlorophylls form concentric helical nanotubes in chlorosomes. Proc Natl Acad Sci U S A 106, 8525–8530 (2009).
Schweizer, H. D. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15, 831–834 (1993).
Vassilieva, E. V. et al. Electron transfer may occur in the chlorosome envelope: the CsmI and CsmJ proteins of chlorosomes are 2Fe–2S ferredoxins. Biochemistry 40, 464–473 (2001).
Vassilieva, E. V. et al. Subcellular localization of chlorosome proteins in Chlorobium tepidum and characterization of three new chlorosome proteins: CsmF, CsmH and CsmX. Biochemistry 41, 4358–4370 (2002).
Acknowledgements
We thank Dr. Yuichiro Kashiyama of Ritsumeikan University for useful discussion. This work was partially supported by Grants-in-Aid for Scientific Research (A) (No. 22245030) (to H.T.) and (C) (No. 24590366 and 24550065) (to M.N. and T.M., respectively) as well as for Young Scientists (B) (No. 24750169) (to J.H.) from the Japanese Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Contributions
J.H., M.N. and H.T. conceived the project. J.H. performed the mutational analyses and bacterial growth measurements. J.H. and T.M. contributed spectroscopic analysis of the bchU mutant cells and its pigments. T.M. determined the compositions of BChl f homologs in the mutant and structure of R[E,E]BChl f. Y.T. isolated chlorosomes from the mutant and measured the UV-Vis-NIR absorption spectra. J.H., Y.T. and H.T analyzed the data, interpreted the results and wrote the manuscript. All authors edited the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Harada, J., Mizoguchi, T., Tsukatani, Y. et al. A seventh bacterial chlorophyll driving a large light-harvesting antenna. Sci Rep 2, 671 (2012). https://doi.org/10.1038/srep00671
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00671
This article is cited by
-
Living off the Sun: chlorophylls, bacteriochlorophylls and rhodopsins
Photosynthetica (2018)
-
Self-aggregation of synthetic zinc methyl 20-substituted 3-hydroxymethyl-pyropheophorbides as models of bacteriochlorophyll-c
Photosynthesis Research (2018)
-
Proposal for probing energy transfer pathway by single-molecule pump-dump experiment
Scientific Reports (2016)
-
In vitro stereospecific hydration activities of the 3-vinyl group of chlorophyll derivatives by BchF and BchV enzymes involved in bacteriochlorophyll c biosynthesis of green sulfur bacteria
Photosynthesis Research (2016)
-
Isolation and structural determination of C8-vinyl-bacteriochlorophyll d from the bciA and bchU double mutant of the green sulfur bacterium Chlorobaculum tepidum
Photosynthesis Research (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.