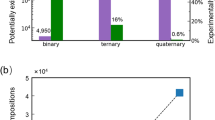
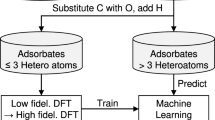
Abstract
The discovery of transition metal complexes (TMCs) with optimal properties requires large ligand libraries and efficient multiobjective optimization algorithms. Here we provide the tmQMg-L library, containing 30k diverse and synthesizable ligands with robustly assigned charges and metal coordination modes. tmQMg-L enabled the generation of 1.37 million palladium TMCs, which were used to develop and benchmark the Pareto-Lighthouse multiobjective genetic algorithm (PL-MOGA). With fine control over aim and scope, this algorithm maximized both the polarizability and highest occupied molecular orbital–lowest unoccupied molecular orbital gap of the TMCs within selected regions of the Pareto front, without requiring prior knowledge on the objective limits. Instead of genetic operations on small ligand fragments, the PL-MOGA did whole-ligand mutation and crossover operations, which in chemical spaces containing billions of systems, yielded thousands of highly diverse TMCs in an interpretable manner.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The tmQMg-L dataset can be accessed at https://github.com/hkneiding/tmQMg-L, including the data for the charge assignment benchmark and the 1.37M space. The dataset is also available via Zenodo at https://doi.org/10.5281/zenodo.10374523 (ref. 64). In addition to the geometric and electronic structure information, it provides Weisfeiler–Lehman graph hashes65. All data are openly available. Source data are provided with this paper. The larger datasets may require Linux software to be visualized.
Code availability
The PL-MOGA code is available from https://github.com/hkneiding/PL-MOGA and Zenodo via https://doi.org/10.5281/zenodo.10663863 (ref. 66), including the DFT geometries of selected TMC hits and the weighted-sum benchmark. The code includes a command line functionality, together with documentation and installation instructions. All code is openly available.
References
Mjos, K. D. & Orvig, C. Metallodrugs in medicinal inorganic chemistry. Chem. Rev. 114, 4540–4563 (2014).
Prier, C. K., Rankic, D. A. & MacMillan, D. W. C. Visible light photoredox catalysis with transition metal complexes: applications in organic synthesis. Chem. Rev. 113, 5322–5363 (2013).
Kalyanasundaram, K. & Gratzel, M. Applications of functionalized transition metal complexes in photonic and optoelectronic devices. Coord. Chem. Rev. 177, 347–414 (1998).
Yoon, T. P., Ischay, M. A. & Du, J. N. Visible light photocatalysis as a greener approach to photochemical synthesis. Nature Chem. 2, 527–532 (2010).
Furukawa, H., Cordova, K. E., O’Keeffe, M. & Yaghi, O. M. The chemistry and applications of metal–organic frameworks. Science 341, 974 (2013).
Balcells, D. & Nova, A. Designing Pd and Ni catalysts for cross-coupling reactions by minimizing off-cycle species. ACS Catal. 8, 3499–3515 (2018).
Foscato, M. & Jensen, V. R. Automated in silico design of homogeneous catalysts. ACS Catal. 10, 2354–2377 (2020).
Robbins, D. W. & Hartwig, J. F. A simple, multidimensional approach to high-throughput discovery of catalytic reactions. Science 333, 1423–1427 (2011).
Nandy, A. et al. Computational discovery of transition-metal complexes: from high-throughput screening to machine learning. Chem. Rev. 121, 9927–10000 (2021).
Huang, B. & von Lilienfeld, O. A. Ab initio machine learning in chemical compound space. Chem. Rev. 121, 10001–10036 (2021).
Freeze, J. G., Kelly, H. R. & Batista, V. S. Search for catalysts by inverse design: artificial intelligence, mountain climbers, and alchemists. Chem. Rev. 119, 6595–6612 (2019).
Kitchin, J. R. Machine learning in catalysis. Nat. Catal. 1, 230–232 (2018).
Gomes, G. D., Pollice, R. & Aspuru-Guzik, A. Navigating through the maze of homogeneous catalyst design with machine learning. Trends Chem. 3, 96–110 (2021).
Friederich, P., Gomes, G. D., De Bin, R., Aspuru-Guzik, A. & Balcells, D. Machine learning dihydrogen activation in the chemical space surrounding Vaska’s complex. Chem. Sci. 11, 4584–4601 (2020).
Nandy, A., Duan, C. R., Goffinet, C. & Kulik, H. J. New strategies for direct methane-to-methanol conversion from active learning exploration of 16 million catalysts. JACS Au 2, 1200–1213 (2022).
Jorner, K., Tomberg, A., Bauer, C., Skold, C. & Norrby, P. O. Organic reactivity from mechanism to machine learning. Nat. Rev. Chem. 5, 240–255 (2021).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Butler, K. T., Davies, D. W., Cartwright, H., Isayev, O. & Walsh, A. Machine learning for molecular and materials science. Nature 559, 547–555 (2018).
Goldberg, D. E. Genetic Algorithms in Search, Optimization, and Machine Learning (Addison-Wesley, 1989).
De Jong, K. A. Evolutionary Computation—A Unified Appraoch (MIT Press, 2006).
Winter, R. et al. Efficient multi-objective molecular optimization in a continuous latent space. Chem. Sci. 10, 8016–8024 (2019).
Anstine, D. M. & Isayev, O. Generative models as an emerging paradigm in the chemical sciences. J. Am. Chem. Soc. 145, 8736–8750 (2023).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: generative models for matter engineering. Science 361, 360–365 (2018).
Le, T. C. & Winkler, D. A. Discovery and optimization of materials using evolutionary approaches. Chem. Rev. 116, 6107–6132 (2016).
Jensen, J. H. A graph-based genetic algorithm and generative model/Monte Carlo tree search for the exploration of chemical space. Chem. Sci. 10, 3567–3572 (2019).
Nigam, A., Pollice, A. & Aspuru-Guzik, A. Parallel tempered genetic algorithm guided by deep neural networks for inverse molecular design. Digit. Discov. 1, 390–404 (2022).
Janet, J. P., Chan, L. & Kulik, H. J. Accelerating chemical discovery with machine learning: simulated evolution of spin crossover complexes with an artificial neural network. J. Phys. Chem. Lett. 9, 1064–1071 (2018).
Gallarati, S., Gerwen, P. V., Schoepfer, A. A., Laplaza, R. & Corminboeuf, C. Genetic algorithms for the discovery of homogeneous catalysts. CHIMIA 77, 39 (2023).
Fey, N., Orpen, A. G. & Harvey, J. N. Building ligand knowledge bases for organometallic chemistry: computational description of phosphorus(III)-donor ligands and the metal-phosphorus bond. Coord. Chem. Rev. 253, 704–722 (2009).
Gugler, S., Janet, J. P. & Kulik, H. J. Enumeration of de novo inorganic complexes for chemical discovery and machine learning. Mol. Syst. Des. Eng. 5, 139–152 (2020).
Gensch, T. et al. A comprehensive discovery platform for organophosphorus ligands for catalysis. J. Am. Chem. Soc. 144, 1205–1217 (2022).
Ioannidis, E. I., Gani, T. Z. H. & Kulik, H. J. molSimplify: a toolkit for automating discovery in inorganic chemistry. J. Comput. Chem. 37, 2106–2117 (2016).
Foscato, M., Venkatraman, V. & Jensen, V. R. DENOPTIM: software for computational de novo design of organic and inorganic molecules. J. Chem. Inf. Model. 59, 4077–4082 (2019).
Sobez, J. G. & Reiher, M. MOLASSEMBLER: molecular graph construction, modification, and conformer generation for inorganic and organic molecules. J. Chem. Inf. Model. 60, 3884–3900 (2020).
Chen, S. et al. Automated construction and optimization combined with machine learning to generate Pt(II) methane C–H activation transition states. Top. Catal. 65, 312–324 (2022).
Kneiding, H. et al. Deep learning metal complex properties with natural quantum graphs. Digit. Discov. 2, 618–633 (2023).
Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. The Cambridge Structural Database. Acta Cryst. B B72, 171–179 (2016).
Duan, C. et al. Exploiting ligand additivity for transferable machine learning of multireference character across known transition metal complex ligands. J. Chem. Theory Comput. 18, 4836–4845 (2022).
Vela, S., Laplaza, R., Cho, Y. R. & Corminboeuf, C. cell2mol: encoding chemistry to interpret crystallographic data. Npj Comput. Mater. 8, 188 (2022).
Matsuoka, W., Harabuchi, Y. & Maeda, S. Virtual ligand-assisted screening strategy to discover enabling ligands for transition metal catalysis. ACS Catal. 12, 3752–3766 (2022).
Gao, W. H. & Coley, C. W. The synthesizability of molecules proposed by generative models. J. Chem. Inf. Model. 60, 5714–5723 (2020).
Chu, Y. H., Heyndrickx, W., Occhipinti, G., Jensen, V. R. & Alsberg, B. K. An evolutionary algorithm for de novo optimization of functional transition metal compounds. J. Am. Chem. Soc. 134, 8885–8895 (2012).
Durrant, M. C. The use of quantum molecular calculations to guide a genetic algorithm: a way to search for new chemistry. Chem. Eur. J. 13, 3406–3413 (2007).
Janet, J. P., Ramesh, S., Duan, C. & Kulik, H. J. Accurate multiobjective design in a space of millions of transition metal complexes with neural-network-driven efficient global optimization. ACS Cent. Sci. 6, 513–524 (2020).
Sowndarya, S. V. S. et al. Multi-objective goal-directed optimization of de novo stable organic radicals for aqueous redox flow batteries. Nat. Mach. Intell. 4, 720–730 (2022).
Verhellen, J. Graph-based molecular Pareto optimisation. Chem. Sci. 13, 7526–7535 (2022).
Hase, F., Roch, L. M. & Aspuru-Guzik, A. Chimera: enabling hierarchy based multi-objective optimization for self-driving laboratories. Chem. Sci. 9, 7642–7655 (2018).
Nigam, A., Pollice, R., Krenn, M., Gomes, G. D. & Aspuru-Guzik, A. Beyond generative models: superfast traversal, optimization, novelty, exploration and discovery (STONED) algorithm for molecules using SELFIES. Chem. Sci. 12, 7079–7090 (2021).
Brown, N., Fiscato, M., Segler, M. H. S. & Vaucher, A. C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 59, 1096–1108 (2019).
Laplaza, R., Gallarati, S. & Corminboeuf, C. Genetic optimization of homogeneous catalysts. Chem. Methods 2, e202100107 (2022).
Seumer, J., Hansen, J. K. S., Nielsen, M. B. & Jensen, J. H. Computational evolution of new catalysts for the Morita–Baylis–Hillman reaction. Angew. Chem. Int. Ed. 62, e202218565 (2023).
Balcells, D. & Skjelstad, B. B. tmQM dataset–quantum geometries and properties of 86k transition metal complexes. J. Chem. Inf. Model. 60, 6135–6146 (2020).
Chen, S. et al. ReaLigands: a ligand library cultivated from experiment and intended for molecular computational catalyst design. J. Chem. Inf. Model. https://doi.org/10.1021/acs.jcim.3c01310 (2023).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 77, 3865–3868 (1996).
Weigend, F. & Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys. Chem. Chem. Phys. 7, 3297–3305 (2005).
von Lilienfeld, O. A., Müller, K. R. & Tkatchenko, A. Exploring chemical compound space with quantum-based machine learning. Nat. Rev. Chem. 4, 347–358 (2020).
Hoffmeister, F. & Sprave, J. Problem-independent handling of constraints by use of metric penalty functions. In Evolutionary Programing (1996); https://ls11-www.cs.tu-dortmund.de/~joe/papers/ep96a.pdf
Devi, R. V., Sathya, S. S. & Coumar, M. S. Multi-objective genetic algorithm for de novo drug design (MoGADdrug). Curr. Comput. Aid. Drug Des. 17, 445–457 (2021).
Pollice, R. et al. Data-driven strategies for accelerated materials design. Acc. Chem. Res. 54, 849–860 (2021).
Hueffel, J. A. et al. Accelerated dinuclear palladium catalyst identification through unsupervised machine learning. Science 374, 1134–1140 (2021).
Adamo, A. & Barone, V. Toward reliable density functional methods without adjustable parameters: the PBE0 model. J. Chem. Phys. 110, 6158–6169 (1999).
Grimme, S., Bannwarth, C. & Shushkov, P. A robust and accurate tight-binding quantum chemical method for structures, vibrational frequencies, and noncovalent interactions of large molecular systems parametrized for all spd-block elements (Z = 1–86). J. Chem. Theory Comput. 13, 1989–2009 (2017).
Bannwarth, C., Ehlert, S. & Grimme, S. GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 15, 1652–1671 (2019).
Kneiding, H., Balcells, D. & Nova, A. tmQMg-L. Zenodo https://doi.org/10.5281/zenodo.10374523 (2023).
Nandy, A., Taylor, M. G. & Kulik, H. J. Identifying underexplored and untapped regions in the chemical space of transition metal complexes. J. Phys. Chem. Lett. 14, 5798–5804 (2023).
Kneiding, H. tmQMg-L. Zenodo https://doi.org/10.5281/zenodo.10663863 (2024).
Acknowledgements
European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement number 945371 (H.K.). This article reflects only the author’s view and the REA is not responsible for any use that may be made of the information it contains. Research Council of Norway (RCN) FRIPRO program supporting the CO2pCat project, with number 314321 (A.N.). RCN FRIPRO program supporting the catLEGOS project, with number 325003 (D.B.). RCN support through the Centers of Excellence program, including the Hylleraas Centre, with project number 262695, and the Sigma2 – National Infrastructure for High Performance Computing and Data Storage in Norway, with grant number NN4654K (H.K., A.N. and D.B.). We also thank M. Strandgaard and T. Linjordet for helpful discussions and for reviewing preliminary versions of this manuscript.
Author information
Authors and Affiliations
Contributions
H.K. was the main developer of the tmQMg-L dataset, the 1.37M space and the PL-MOGA algorithm. H.K. also derived the combinatorics of the square planar TMC space and developed the concept of a generative model based on whole-ligand multiple-site genetic operations. A.N. and D.B. developed the concept of extracting the ligand charges from the natural Lewis structures. All authors made substantial contributions to the conception and design of the work. D.B. was the main contributor to the writing and revision of the manuscript, as well as to the definition, supervision and funding of the research project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Jan Jensen, Aditya Nandy and Robert Pollice for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary sections, Figs. 1–20, Equations 1–4, Algorithm 1 and Table 1.
Source data
Source Data Fig. 3
Data plotted in Fig. 3a,b, in .csv format.
Source Data Fig. 4
Data plotted in Fig. 4a–d, in .csv format.
Source Data Fig. 5
Data plotted in Fig. 5a,b, in .csv format.
Source Data Fig. 6
Data plotted in Fig. 6a, in .csv format.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kneiding, H., Nova, A. & Balcells, D. Directional multiobjective optimization of metal complexes at the billion-system scale. Nat Comput Sci 4, 263–273 (2024). https://doi.org/10.1038/s43588-024-00616-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-024-00616-5