Abstract
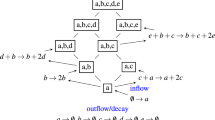
Modern applications involving multispecies microbial communities rely on the ability to predict structures of such communities in defined environments. The structures depend on pairwise and high-order interactions between species. To unravel these interactions, classical bottom-up approaches examine all possible species subcommunities. Such approaches are not scalable as the number of subcommunities grows exponentially with the number of species, n. Here we present a top-down method wherein the number of subcommunities to be examined grows linearly with n, drastically reducing experimental effort. The method uses steady-state data from leave-one-out subcommunities and mathematical modeling to infer effective pairwise interactions and predict community structures. The accuracy of the method increases with n, making it suitable for large communities. We established the method in silico and validated it against a five-species community from literature and an eight-species community cultured in vitro. Our method offers an efficient and scalable tool for predicting microbial community structures.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source Data are provided with this paper.
Code availability
The computer code used to implement EPICS is available on GitHub (https://github.com/narendradixit/EPICS/tree/v1) and on Zenodo62.
References
Baldrian, P. Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol. Rev. 41, 109–130 (2017).
Jansson, J. K. & Hofmockel, K. S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 18, 35–46 (2020).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Cani, P. D. Human gut microbiome: hopes, threats and promises. Gut 67, 1716–1725 (2018).
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Lamont, R. J., Koo, H. & Hajishengallis, G. The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759 (2018).
Valdes, A. M., Walter, J., Segal, E. & Spector, T. D. Role of the gut microbiota in nutrition and health. Brit. Med. J. 361, k2179 (2018).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Valles-Colomer, M. et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 4, 623–632 (2019).
Angulo, M. T., Moog, C. H. & Liu, Y.-Y. A theoretical framework for controlling complex microbial communities. Nat. Commun. 10, 1045 (2019).
Fuhrman, J. A. Microbial community structure and its functional implications. Nature 459, 193–199 (2009).
Inda, M. E., Broset, E., Lu, T. K. & de la Fuente-Nunez, C. Emerging frontiers in microbiome engineering. Trends Immunol. 40, 952–973 (2019).
Prosser, J. I. et al. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392 (2007).
Sheth, R. U., Cabral, V., Chen, S. P. & Wang, H. H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200 (2016).
Xiao, Y., Angulo, M. T., Lao, S., Weiss, S. T. & Liu, Y.-Y. An ecological framework to understand the efficacy of fecal microbiota transplantation. Nat. Commun. 11, 3329 (2020).
Bittleston, L. S., Gralka, M., Leventhal, G. E., Mizrahi, I. & Cordero, O. X. Context-dependent dynamics lead to the assembly of functionally distinct microbial communities. Nat. Commun. 11, 1440 (2020).
Lawson, C. E. et al. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741 (2019).
Aroniadis, O. C. & Brandt, L. J. Fecal microbiota transplantation: past, present and future. Curr. Opin. Gastroenterol. 29, 79–84 (2013).
Chng, K. R. et al. Metagenome-wide association analysis identifies microbial determinants of post-antibiotic ecological recovery in the gut. Nat. Ecol. Evol. 4, 1256–1267 (2020).
Zhou, K., Qiao, K., Edgar, S. & Stephanopoulos, G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 33, 377–383 (2015).
Mee, M. T., Collins, J. J., Church, G. M. & Wang, H. H. Syntrophic exchange in synthetic microbial communities. Proc. Natl Acad. Sci. USA 111, E2149–E2156 (2014).
Mukherjee, S. & Bassler, B. L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382 (2019).
Scott, S. R. & Hasty, J. Quorum sensing communication modules for microbial consortia. ACS Synth. Biol. 5, 969–977 (2016).
Jagmann, N. & Philipp, B. Design of synthetic microbial communities for biotechnological production processes. J. Biotechnol. 184, 209–218 (2014).
McCarty, N. S. & Ledesma-Amaro, R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 37, 181–197 (2019).
Vázquez-Castellanos, J. F., Biclot, A., Vrancken, G., Huys, G. R. & Raes, J. Design of synthetic microbial consortia for gut microbiota modulation. Curr. Opin. Pharmacol. 49, 52–59 (2019).
Meyer, A. et al. Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors. Nat. Chem. 7, 673–678 (2015).
Widder, S. et al. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10, 2557–2568 (2016).
Friedman, J., Higgins, L. M. & Gore, J. Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 0109 (2017).
Goldford, J. E. et al. Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018).
Xiao, Y. et al. Mapping the ecological networks of microbial communities. Nat. Commun. 8, 2042 (2017).
Machado, D. et al. Polarization of microbial communities between competitive and cooperative metabolism. Nat. Ecol. Evol. 5, 195–203 (2021).
Pacheco, A. R., Moel, M. & Segrè, D. Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. Commun. 10, 103–103 (2019).
Pacheco, A. R., Osborne, M. L. & Segrè, D. Non-additive microbial community responses to environmental complexity. Nat. Commun. 12, 2365 (2021).
Gould, A. L. et al. Microbiome interactions shape host fitness. Proc. Natl Acad. Sci. USA 115, E11951–E11960 (2018).
Ansari, A. F. et al. High-order interactions can eclipse pairwise interactions in shaping the structure of microbial communities. Ind. Eng. Chem. Res. 58, 23508–23518 (2019).
Bairey, E., Kelsic, E. D. & Kishony, R. High-order species interactions shape ecosystem diversity. Nat. Commun. 7, 12285 (2016).
Grilli, J., Barabás, G., Michalska-Smith, M. J. & Allesina, S. Higher-order interactions stabilize dynamics in competitive network models. Nature 548, 210–213 (2017).
Biggs, M. B., Medlock, G. L., Kolling, G. L. & Papin, J. A. Metabolic network modeling of microbial communities. WIRES Syst. Biol. Med. 7, 317–334 (2015).
Harcombe, W. R. et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 7, 1104–1115 (2014).
Levy, R. & Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl Acad. Sci. USA 110, 12804–12809 (2013).
Ravikrishnan, A. & Raman, K. Systems-Level Modelling of Microbial Communities (CRC, 2018); https://doi.org/10.1201/9780429487484
Cammarota, G. et al. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 635–648 (2020).
Faust, K. & Raes, J. Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550 (2012).
Guo, X. & Boedicker, J. Q. The contribution of high-order metabolic interactions to the global activity of a four-species microbial community. PLoS Comput. Biol. 12, e1005079 (2016).
Sanchez-Gorostiaga, A., Bajić, D., Osborne, M., Poyatos, J. & Sanchez, A. High-order interactions distort the functional landscape of microbial consortia. PLoS Biol. 17, e3000550 (2019).
Leventhal, G. E. et al. Strain-level diversity drives alternative community types in millimetre-scale granular biofilms. Nat. Microbiol. 3, 1295–1303 (2018).
Bosi, E., Bacci, G., Mengoni, A. & Fondi, M. Perspectives and challenges in microbial communities metabolic modeling. Front. Genet. 8, 88 (2017).
Babaei, P., Shoaie, S., Ji, B. & Nielsen, J. Challenges in modeling the human gut microbiome. Nat. Biotechnol. 36, 682–686 (2018).
Großkopf, T. & Soyer, O. S. Synthetic microbial communities. Curr. Opin. Microbiol. 18, 72–77 (2014).
Johns, N. I., Blazejewski, T., Gomes, A. L. C. & Wang, H. H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 31, 146–153 (2016).
De Roy, K., Marzorati, M., Van den Abbeele, P., Van de Wiele, T. & Boon, N. Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities. Environ. Microbiol. 16, 1472–1481 (2014).
Venturelli, O. S. et al. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol. 14, e8157 (2018).
Zomorrodi, A. R. & Segrè, D. Synthetic ecology of microbes: mathematical models and applications. J. Mol. Biol. 428, 837–861 (2016).
Kuntal, B. K., Gadgil, C. & Mande, S. S. Web-gLV: a web based platform for Lotka–Volterra based modeling and simulation of microbial populations. Front. Microbiol. 10, 288 (2019).
Marino, S., Baxter, N. T., Huffnagle, G. B., Petrosino, J. F. & Schloss, P. D. Mathematical modeling of primary succession of murine intestinal microbiota. Proc. Natl Acad. Sci. USA 111, 439–444 (2014).
Vos, M. G. J. de, Zagorski, M., McNally, A. & Bollenbach, T. Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc. Natl Acad. Sci. USA 114, 10666–10671 (2017).
Opper, M. & Diederich, S. Replicator dynamics. Comput. Phys. Commun. 121, 141–144 (1999).
Bolnick, D. I. et al. Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol. Lett. 17, 979–987 (2014).
Momeni, B., Xie, L. & Shou, W. Lotka–Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. eLife 6, e25051 (2017).
Ansari, A. F., Reddy, Y. B. S., Raut, J. & Dixit. N. M. EPICS Version v1 (Zenodo, 2021); https://doi.org/10.5281/zenodo.5156236
Acknowledgements
We thank K. Raman and S. Jhunjhunwala for insightful comments.
Author information
Authors and Affiliations
Contributions
A.F.A. and N.M.D. developed the theory. A.F.A. performed the numerical simulations and analyzed the empirical data. Y.B.S.R. and J.R. performed the experiments. All authors interpreted the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Computational Science thanks Boyang Ji and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Editor recognition statement Handling editor: Ananya Rastogi, in collaboration with the Nature Computational Science team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Consistency of effective pairwise interactions from leave-one-out and leave-two-out subcommunities.
The RMS relative error between the interactions estimated using n − 1 species subcommunities and n − 2 species subcommunities for different n using parameter settings corresponding to the first four points in the 3D diagonal in Fig. 3A, indicated as 1, 2, 3, and 4 above.
Extended Data Fig. 2 Exactness of EPICS with purely pairwise interactions.
Comparisons between effective pairwise interactions estimated using EPICS and true pairwise interactions underlying in silico 5-species communities following the GLV model (see Methods). The interaction strengths are depicted in the panels. (A) Pairwise interactions alone are present. (B) Pairwise and ternary interactions present; (C) Pairwise and quaternary interactions present; (D) All three interaction types present. Each dot represents one pairwise interaction in one realization. The red dashed line represents y=x.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, and Figs. 1–15.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Rights and permissions
About this article
Cite this article
Ansari, A.F., Reddy, Y.B.S., Raut, J. et al. An efficient and scalable top-down method for predicting structures of microbial communities. Nat Comput Sci 1, 619–628 (2021). https://doi.org/10.1038/s43588-021-00131-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-021-00131-x
This article is cited by
-
Nutritional and host environments determine community ecology and keystone species in a synthetic gut bacterial community
Nature Communications (2023)
-
Autoencoder neural networks enable low dimensional structure analyses of microbial growth dynamics
Nature Communications (2023)
-
Steering and controlling evolution — from bioengineering to fighting pathogens
Nature Reviews Genetics (2023)
-
Microbial community dynamics revisited
Nature Computational Science (2021)