Abstract
Nitrogen (N) is an essential element for plant growth, and its deficiency influences plants at several physiological and gene expression levels. Barley (Hordeum vulgare) is one of the most important food grains from the Poaceae family and one of the most important staple food crops. However, the seed yield is limited by a number of stresses, the most important of which is the insufficient use of N. Thus, there is a need to develop N-use effective cultivars. In this study, comparative physiological and molecular analyses were performed using leaf and root tissues from 10 locally grown barley cultivars. The expression levels of nitrate transporters, HvNRT2 genes, were analyzed in the leaf and root tissues of N-deficient (ND) treatments of barley cultivars after 7 and 14 days following ND treatment as compared to the normal condition. Based on the correlation between the traits, root length (RL) had a positive and highly significant correlation with fresh leaf weight (FLW) and ascorbate peroxidase (APX) concentration in roots, indicating a direct root and leaf relationship with the plant development under ND. From the physiological aspects, ND enhanced carotenoids, chlorophylls a/b (Chla/b), total chlorophyll (TCH), leaf antioxidant enzymes such as ascorbate peroxidase (APX), peroxidase (POD), and catalase (CAT), and root antioxidant enzymes (APX and POD) in the Sahra cultivar. The expression levels of HvNRT2.1, HvNRT2.2, and HvNRT2.4 genes were up-regulated under ND conditions. For the morphological traits, ND maintained root dry weight among the cultivars, except for Sahra. Among the studied cultivars, Sahra responded well to ND stress, making it a suitable candidate for barely improvement programs. These findings may help to better understand the mechanism of ND tolerance and thus lead to the development of cultivars with improved nitrogen use efficiency (NUE) in barley.
Similar content being viewed by others
Introduction
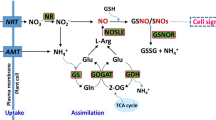
Nitrogen (N) is one of the most important components of biomolecules, amino acids, nucleotides, proteins, chlorophyll, and many plant hormones, as well as an essential component for plant growth and development. Regulating leaf photosynthesis is a crucial role of nitrogen (N) as a mineral nutrient1. The global production volume of barley amounted to about 150.48 million metric tons in the 2022/2023 cropping season year, increasing from around 145.93 million metric tons in 2021/20222. N deficiency (ND) negatively affects leaf chlorosis, bud growth, and overall plant growth3. The deficiency of this essential element causes nutrient imbalance affecting several metabolic pathways including the increased production of reactive oxygen species (ROS)4,5. In rice, ND and the antioxidant system resulted in decreased light-harvesting capacity and increased thermal dissipation of absorbed energy6. An increase in ROS imposes oxidative stress on plants, which utilize antioxidant enzymes, such as ascorbate oxidase (APX), peroxidases (POX), and catalase (CAT), to prevent excessive ROS accumulation7. Wheat genotypes responded differentially to N supply in relation to leaf growth and photosynthesis as well as the maintenance of metabolic constituents8. At the early vegetative stage of plant life, ND adversely influences crop yield, which cannot be offset by N application at later stages9. Nitrogen fertilizer is applied to enhance crop yield because its availability strongly affects crop productivity. A significant amount of N contaminates ground and surface water and emits the greenhouse gas, nitrous oxide10. Thus, crop genotype development with improved nitrogen use efficiency (NUE) can aid in sustainable agriculture and high productivity under low-input conditions. NUE is a complex trait involving physiological, developmental, and environmental factors. This includes the plant's ability to absorb, transport, and remobilize N in the soil. Considerable efforts have been made to understand the molecular basis of plant response to N and to detect N-responsive genes due to NUE complexity11.
Previously, relative comparisons were made for N deficiency tolerance (NDT) traits (shoot and root biomass, plant height, root length, and chlorophyll content) under low N and control conditions. These traits were considered the selection criteria for identifying rice genotypes with improved adaptability12. N uptake by plants is mainly associated with nitrate transporters (NRTs). For instance, of seven NRT2s identified in Arabidopsis thaliana13, AtNRT2.1, AtNRT2.2, AtNRT2.4, and AtNRT2.5 are responsible for approximately 95% of total NO-3 uptake under N-limited conditions14,15. Four NRT2 genes were found in rice16. To provide N sources, high-tension transporters NRT2 and NRT3 play important roles in N uptake13. The role of NRT2.1 in the high-affinity NO− 3 (HATs) transport system has been proved in some studies16,17. The NRT2 transporters have a high affinity for NO-3 and are induced under NO-3-limiting conditions18. However, a major limitation is only using a single genotype in most of the studies. Barley, a robust cereal crop grown in a wide range of agricultural settings from highly developed to subsistence environments, ranks fourth in global importance behind wheat, maize, and rice19. To develop N-effective barely genotypes, it is necessary to identify candidate genes having a critical role in NUE. In the present study, traits were investigated in 10 different varieties of barley to identify candidate genes and molecular mechanisms involved in N. In addition, the physiological and morphological traits of these cultivars were evaluated in response to ND stress. The expression of genes related to NO-3 metabolism in leaves and roots was analyzed under NO-3-limiting conditions to highlight the correlation between the traits and gene expression related to N metabolism.
Results
Morphological traits under ND conditions
Among the morphological traits measured 7 and 14 days after applying ND, the Sahra cultivar showed a significant increase in dry root weight (DRW) 7 days after ND application as compared to normal conditions. However, the Noroz cultivar showed a significant decrease in DRW 7 and 14 days after ND application as compared to normal conditions. After 14 days of ND application, Noroz showed a significant decrease in fresh root weight (FRW) (Table 1).
Physiological traits under ND conditions
Chlorophylls a/b
The Sahra and Yousef cultivars showed a significant increase in the chlorophyll a (Ch-a) content 7 and 14 days after the ND application (Fig. 1a). Ch-b content increased significantly in the Sahra and Yousef cultivars 7 and 14 days after the ND application. When compared with normal conditions, the Armaghan cultivar showed a significant increase in Ch-b only 7 days after the ND application (Fig. 1b).
The effects of ND on 10 barley cultivars for chlorophyll a (a), chlorophyll b (b), total chlorophyll (c), carotenoid (d), and protein content (e) at 7 and 14 days stress periods. Values represent means of three replications per treatment. Different letters demonstrate significant differences between treatments (P < 0.05, Duncan’s Multiple Range Test). NC7:7 days after ND application; NC14: 14 days after ND application.
Total chlorophyll content
Total chlorophyll (TCH) content increased significantly in the Sahra and Yousef cultivars 7 and 14 days after the ND application as compared to normal conditions. The Nimroz cultivar showed a significant decrease in TCH content at both time points after the ND application. In the Goharan cultivar, a significant increase in TCH content was observed only 7 days after the ND application (Fig. 1c).
Carotenoid content
The Khtam cultivar showed a significant decrease in carotenoid content 7 days after the ND application whereas the Nimroz cultivar showed a significant increase in carotenoid content when compared to normal conditions 14 days after the ND application (Fig. 1d).
Protein content
The Zehak, Nobahar, Khatam, and Oxin cultivars showed significant increases in protein content 7 and 14 days after the ND application. As compared to normal conditions, the Sahra cultivar showed a significant increase in protein content 14 days after the ND application. The Nimroz cultivar showed a significant decrease in protein content 7 and 14 days after the ND application as compared to normal treatment (Fig. 1e).
APX content in leaves
The analysis of the APX enzyme in the leaves of Sahra and Goharan cultivars showed a significant increase 14 days after the ND application. The Nobahar, Oxin, and Zehak cultivars showed a significant increase 7 and 14 days after applying ND compared to the normal treatment (Fig. 2a).
The effects of ND on APX (a), CAT (b), and POD (c) antioxidative enzyme activities in shoot of barley cultivars under different concentrations. The error bars (mean ± SE) followed by various letters are statistically significant (P < 0.05, Duncan’s Multiple Range Test). Significant differences between the two concentrations are marked with different letters. NC7:7 days after ND application; NC14: 14 days after ND application.
CAT content in leaves
The Yousef, Oxin, Khatm, and Goharan cultivars showed significant increases in the CAT content of leaves 7 and 14 days after the ND application as compared to the normal treatment. The CAT content increased significantly in the Sahra cultivar 7 days after the ND application. Further, a significant decrease occurred in the CAT content 14 days after the ND application while a significant increase was observed in the Noroz cultivar 7 days after the ND application (Fig. 2b).
POD content in leaves
The POD enzyme showed significant differences in response to the ND application in different cultivars at the two times. The Yousef, Nimroz, Nobahar, Zahak, and Sahra cultivars showed significant increases in the POD content 7 and 14 days after the ND application as compared to the normal treatment. After 14 days of the ND application, POD content significantly increased in Noroz and Goharan cultivars as compared to the normal treatment. In the Armaghan cultivar, a significant decrease was observed 7 and 14 days after the ND application (Fig. 2c).
APX content in roots
The analysis of physiological traits in response to ND showed that the APX enzyme significantly varied in the roots 7 and 14 days after the ND application. The Sahra, Yousef, Nobahar, Zehak, and Nimroz cultivars showed significant increases in the APX content 7 and 14 days after the ND application (Fig. 3a).
The effects of ND on APX (a), CAT (b), and POD (c) antioxidative enzyme activities in root of barley cultivars under different concentrations. The error bars (mean ± SE) followed by various letters are statistically significant (P < 0.05, Duncan’s Multiple Range Test). Significant differences between the two concentrations are marked with different letter. NC7:7 days after ND application; NC14: 14 days after ND application.
CAT content in roots
The analysis of CAT content in response to ND showed that Yousef, Sahra, and Nimroz cultivars were not significantly different between the two ND application times. In the Oxin cultivar, a significant increase in the CAT content was observed 7 days after the ND application. In the Nobahar cultivar, CAT content increased significantly 7 and 14 days after the ND application. Zehak and Goharan showed a significant decrease 7 days after the ND application (Fig. 3b).
POD content in roots
The analysis of the POD enzyme in response to ND showed significant differences among 10 barley cultivars. The Sahra, Nimroz, Noroz, Oxin, Yousef, Khatam, and Nobahar cultivars showed a significant increase in the POD content 7 days after the ND application. Armaghan showed a significant decrease in POD content 14 days after the ND application (Fig. 3c).
The HvNRT2 gene expression profile in shoots in response to ND
The expression of 10 genes in response to ND showed that the most expressed genes in shoots belonged to Sahra, Zehak, and Yousef cultivars 7 and 14 days after the ND application. In the Sahra cultivar, all HvNRT2 genes were significantly increased 7 days after applying ND, as compared to 14 days after the ND application. In the Zehak cultivar, the HvNRT2.1, HvNRT2.2, HvNRT2.3, HvNRT2.4, HvNRT2.5, HvNRT2.7, and HvNRT2.10 genes showed a higher significant expression after 7 days than 14 days after the ND application. In the Yousef cultivar, the HvNRT2.1, HvNRT2.2, HvNRT2.3, HvNRT2.4, HvNRT2.5, HvNRT2.8, HvNRT2.9, and HvNRT2.11 genes revealed a higher significant expression 7 days after the ND application than 14 days after this treatment (Fig. 4). The Noroz cultivar showed an up-regulation of the HvNRT2.5 gene 14 days after the ND application. In the Oxin cultivar, the HvNRT2.2, HvNRT2.5, and HvNRT2.7 genes revealed a higher significant expression 7 days after applying ND than after 14 days. After 14 days of applying ND, the expression levels of HvNRT2.8 and HvNRT2.9 genes increased as compared to 7 days after the ND application. The 14-day ND application led to an increase in the expression levels of HvNRT2.4 and HvNRT2.5 genes in the Nobahar cultivar.
In the Nimroz cultivar, the HvNRT2.2, HvNRT2.3, HvNRT2.9, and HvNRT2.11 genes showed a significantly higher expression 7 days after the ND application than after 14 days. The expression of HvNRT2.5 and HvNRT2.7 genes increased in the Nimroz cultivar after 14 days as compared to 7 days after the ND application (Fig. 4). In the Khatam cultivar, the HvNRT2.4 and HvNRT2.9 genes were significantly expressed after 7 days as compared to the 14-day ND application. The expression of HvNRT2.3, HvNRT2.7, and HvNRT2.8 genes indicated significantly higher levels 14 days after applying the ND than after 7 days. In the Goharan cultivar, the HvNRT2.6 gene showed a significantly higher expression 7 days after applying ND than after 14 days. A significantly higher expression was observed in the HvNRT2.1 gene 14 days after the ND application than after 7 days. In the Armaghan cultivar, the HvNRT2.7 gene showed a significantly higher expression after 14 days than the 7-day ND application (Fig. 4).
The HvNRT2 gene expression profile in roots in response to ND
The expression profile of the 10 nitrate-transporter genes showed that HvNRT2.6 and HvNRT2.11 genes were expressed significantly in the Armaghan cultivar 7 days after applying ND as compared to after 14 days. After 14 days of applying ND, the HvNRT2.1, HvNRT2.3, HvNRT2.4, HvNRT2.7, HvNRT2.8, and HvNRT2.9 genes showed a significantly higher expression than 7 days after the ND application (Fig. 6). In the Goharan cultivar, the expression of HvNRT2.1, HvNRT2.2, HvNRT2.7, HvNRT2.9, and HvNRT2.11 genes increased significantly 7 days after applying ND. The expression of HvNRT2.4, HvNRT2.5, HvNRT2.8, HvNRT2.9, and HvNRT2.11 genes significantly increased in the Khatam cultivar 7 days after applying ND (Fig. 5). After 14 days of applying ND, the HvNRT2.1 gene expression showed a significant increase as compared to 7 days after applying ND. In the Nimroz cultivar, the HvNRT2.4 gene expression significantly increased after 7 days as compared to 14 days of applying ND. Two genes, HvNRT2.8 and HvNRT2.9, were significantly expressed after 14 days as compared to 7 days after the ND application. In the Nobahar cultivar, the expression of HvNRT2.3 and HvNRT2.4 genes rose significantly 7 days after applying ND. After 14 days of applying ND, the HvNRT2.6 gene showed a significant increase in expression (Fig. 6). In the Noroz cultivar, significant increases in the HvNRT2.1 and HvNRT2.2 gene expression were observed 14 days after the ND application. The expression of HvNRT2.7, HvNRT2.8, and HvNRT2.9 genes was redoubled significantly in the Oxin cultivar 14 days after the ND application. In the Sahra cultivar, all HvNRT2 genes showed a highly significant elevated expression after 14 days as compared to 7 days after the ND application (Fig. 5). In the Yousef cultivar, significantly increased expression levels of the HvNRT2.1, HvNRT2.2, HvNRT2.3, HvNRT2.5, and HvNRT2.8 genes occurred 7 days after the ND application. After 14 days of applying ND, only the HvNRT2.6 gene showed an increase in expression as compared to 7 days after this treatment. In the Zehak cultivar, the expression of HvNRT2.4, HvNRT2.6, HvNRT2.7, HvNRT2.8, HvNRT2.9, and HvNRT2.11 genes rose significantly after 7 days as compared to 14 days after the ND application (Fig. 5).
The correlation coefficients of physiological and morphological traits and gene expression at 7 (a) and 14 (b) days after ND application. Protein content (PC); plant height (PH); leaf dry weight (LDW); leaf fresh weight (LFW); root fresh weight (RFW); root dry weight (RDW); HvNRT2r (root HvNRT2); HvNRT2l (leaf HvNRT2).
Correlations between the measured traits and gene expression levels 7 and 14 days after the ND condition
Correlations between traits 7 days after ND conditions
The HvNRT2.7l had a positive correlation with HvNRT2.8r. The HvNRT2.7l showed a negative correlation with carotenoid content whereas the HvNRT2.4r was positively correlated with HvNRT2.9r and HvNRT2.11r. The Ch-b and TCH had a positive correlation with FRW. The HvNRT2.9r and HvNRT2.11r showed a negative correlation with FRW, and carotenoid content was negatively correlated with Ch-a content (Fig. 6a).
Correlations between the traits 14 days after ND
The RL had positive and highly significant correlations with FLW and APX in roots. The FLW showed positive and significant correlations with APX, POD, CAT, FRW, and DLW in roots. The plant height (PH) had a positive and significant correlation with the root POD. The root APX had a positive and significant correlation with the root POD. The leaf CAT and HvNRT2.1l were positively correlated with leaf POD and HvNRT2.8l, respectively. The HvNRT2.11l was positively correlated with HvNRT2.9l, HvNRT2.2l, and HvNRT2.8l. Positive correlations were observed between HvNRT2.2l and HvNRT2.5r, HvNRT2.9l, and HvNRT2.8l. The HvNRT2.7r was positively correlated with HvNRT2.9r and HvNRT2.8r. The HvNRT2.9r had positive correlations with HvNRT2.8r and HvNRT2.3l. The HvNRT2.6l showed a positive correlation with PC, and PC was positively correlated with HvNRT2.4l. There was a positive correlation between HvNRT2.4l and HvNRT2.6r (Fig. 6b).
Cluster analysis of the cultivars in response to ND
The cluster analysis of morphological and physiological traits showed that the cultivars were divided into four groups. Tolerant and semi-tolerant cultivars, such as Zehak (NC7 and NC14), Oxin (NC7 and NC14), Sahra (NC7), and Yousef (NC7 and NC14), were placed in the first cluster. The Nobahar (NC7), a sensitive cultivar, was placed in the second cluster. The third cluster included semi-tolerant cultivars, such as Armaghan (NC7 and NC14), Sahra (NC14), Nimroz (NC7), and Noroz (NC14) (Fig. 7a). The fourth cluster included Khatam (NC7 and NC14), Goharan (NC7 and NC14), Nimroz (NC14), Noroz (NC7), and Nobahar (NC14) as sensitive and semi-sensitive cultivars. The diversity of cultivars in all environments for morphological and physiological traits was evaluated using the bi-plot graphic display, and the cultivars were evaluated according to their principal component analysis (PCA) scores. The two principle components determined 80.7% of the variation. The Noroz, Yousef, Oxin, Zehak, and Sahra cultivars are considered tolerant cultivars located on the upper right side of the graph. The Khatam, Nimroz, Nobahar, Armaghan, and Nobahar cultivars are considered sensitive and semi-sensitive cultivars located on the left bottom of the diagram (Fig. 7b).
Discussion
Nitrogen is one of the main components of amino acids, proteins, and nucleic acids, which are core nutrients for the building blocks of plant cells20. Cultivars that use N more efficiently are one of the main objectives of barley breeding programs. The physiological, morphological, and transcriptional analyses were incorporated in this study to obtain insights into the growth of 10 barely cultivars on ND. Sahra revealed a high DRW, protein content, Chl a/b, TCH, APX, CAT, and POD in roots and leaves after ND treatment. Furthermore, the highest HvNRT2 gene expression was measured in Sahra after the ND application. In the present study, the 10 barely cultivars were significantly different for DRW and leaf chlorophyll content at the seedling stage under ND, suggesting that those two indicators could be used for screening barely cultivars under ND conditions at the seedling stage.
Plant growth and development traits
Significant differences among the 10 cultivars were detected for FRW, DRW, and FLW. The differences in growth performance and N accumulation between the 10 barely cultivars indicated that Sahra and Yousef could maintain better growth than the other eight cultivars under the LN treatment. The DRW was higher in Sahra than in the other nine cultivars. Based on the previous studies, root development might be related to the ND-induced signaling cascade21,22. Root growth was stimulated by both the increased root uptake area and reduced nutrient demand in shoots23. In C. odorata plants experiencing nitrogen deficiency, metabolic resources were allocated preferentially to root system growth24.
Physiological traits in ND application
On 7 and 14 days after the ND application, significant increases in Chl-a/b, TCH, and antioxidant enzyme content in Sahra, Yousef, and Zehak cultivars were observed as compared to normal treatment; therefore, these are considered ND-tolerant cultivars. An indicator of plant growth and development is photosynthetic capacity25. In our study, Sahra, Yousef, and Zehak increased photosynthetic efficiency compared to seven cultivars, indicating a high tolerance to ND. Chlorophylls a/b in leaves were increased in tolerant cultivars (Sahra and Yousef) 7 and 14 days after the application of ND. According to other findings, leaf chlorophylls a/b were increased under ND in wheat26 and rice27. In previous studies, significant differences in chlorophyll concentrations were found in sorghum, rice, maize, and pearl millet under ND conditions28. The main ROS-scavenging enzymes in plants are POD, APX, and CAT. The observed increases in the activities of these enzymes contribute to an increase in AOX activity, especially in plants subjected to ND29.
Gene expression analyses 7 and 14 days after ND conditions
To identify the molecular mechanisms adopted by ND-tolerant barley cultivars under ND conditions, the differences in the expression levels of HvNRT2 genes involved in nitrogen metabolism were compared between ND-tolerant and ND-sensitive barley cultivars. On 7 and 14 days after ND, Sahra, Zehak, and Yousef showed higher significant expression levels in leaves and roots than the other cultivars. At the seedling stage, HvNRT2.1, HvNRT2.2, HvNRT2.4, and HvNRT2.5 genes were mainly expressed in leaves and roots 7 days after the ND application. The AtNRT2.1, AtNRT2.2, AtNRT2.4, and AtNRT2.5 play key roles in root NO-3 uptake in Arabidopsis30. The potential role of NRT2.1 as a nitrate detector has been suggested due to its strong attraction toward nitrate and its influence on root structure in environments with limited nitrogen31. In cotton, the overexpression of NRT2 genes could increase both nitrate uptake and transport as well as the plant biomass in response to ND32. The presence of NRT2 genes is crucial in enhancing the uptake and transport of N in conditions where it is deficient. NRT2.4 is believed to facilitate the transportation of N from roots to shoots in response to N deficiency33. In addition, while NRT2.4 and NRT2.5 genes were predominantly expressed in roots, their presence was also observed in shoots, indicating that these genes play a role in N transport during nitrogen deficiency34.
Correlations and cluster analyses of the 10 cultivars on ND conditions
Based on the data from the 10 barely cultivars and the correlation analysis, the modifications of FLW were closely correlated with DLW, RL, and FRW, demonstrating that root and shoot development could also be considered useful indicators for the evaluation of the plant’s ND response. In our study, the Sahra possessed high FLW, DLW, and FRW as compared with the other cultivars. Cultivars such as Sahra and Yousef showed a significant increase in leaf CAT content. In the tolerant genotypes, antioxidant enzymes showed a significant increase, indicating the high tolerance of these cultivars in response to ND stress in the long term. According to our findings, tolerant cultivars had greater antioxidant enzyme activity in the root and leaf than sensitive accessions. A positive correlation between NUE and root development was observed in rice under low and high nitrogen levels, indicating a relationship between NUE and root growth35. The results of the PCA and cluster analysis showed that tolerant and sensitive cultivars were placed in a group with almost a similar pattern under ND stress. The Sahra, Yousef, and Zehak cultivars were nitrogen-tolerant cultivars.
Conclusion
In the present study, 10 cultivars were exposed to ND application for 7 and 14 days. The HvNRT2 genes were up-regulated suggesting that the nutrient transporting and antioxidant regulation may play an important role in barely in response to ND. A positive correlation between NUE and root development was observed under ND. Overall, Sahra cultivar showed high biomass and protein content in response to ND stress, making it a good candidate as a tolerant ND cultivar at seedling stage.
Methods
The plant material was officially obtained from the Seed and Plant Improvement Institute (SPII), formally identified by the corresponding author Dr. Abbas Saidi, and confirmed by Dr. Habibollah Ghazvini in the Cereal Research Department (CRD), at the SPII, which deposits the collected material. Furthermore, the use of plants in the present study complies with all necessary international, national, and institutional guidelines and legislation. All plant material is owned by the authors and no permission is required for their use. Information on the voucher specimen was obtained by Dr. Habibollah Ghazvini. The seeds of 10 barley cultivars were germinated on wet Whatman filter papers in Petri dishes (Table 2). Seven to 10 day-old seedlings with a uniform growth state, were moved to 10-L containers. The plants were treated with a modified Hoagland nutrient solution36 containing 2 mmol/L of NH+4NO-3, 0.4 mmol/L of MgSO4, 0.3 mmol/L of K2SO4, 0.2 mmol/L of KH2PO4, 0.4 mmol/L of CaCl2, 0.19 µmol/L of CuSO4, 46.9 µmol/L of H3BO3, 4.5 µmol/L of MnCl2, 1 µmol/ L of Na2MoO4, 0.38 µmol/L of ZnSO4, and 19.9 µmol/L of Fe (III) EDTA. The pH of the solution was adjusted to 5.8 with NaOH. The two N treatments included 0.2 mmol/L of NH+4NO-3 (ND) and 2 mmol/L NH+4NO-3 (as a control). Plant height (PH), dry leaf weight (DLW), dry root weight (DRW), fresh leaf weight (FLW), fresh root weight (FRW), and root length (RL) were recorded after 7 and 14 days of N treatments. Plants with a uniform growth status were subsequently harvested as replicates, separated into roots and shoots, and dried in an oven at 72 °C for 3 days to obtain DRW and DLW.
Characterization of physiological traits
To investigate the physiological traits, normal and stressed barley leaves and roots were collected at the two-week seedling stage 7 and 14 days after the application ND. Fresh leaf samples were washed with distilled water in the laboratory and then left to dry at room temperature (18 °C) for 6 h for the analysis of chlorophylls (Chl-a and Chl-b) and carotenoid contents. An accurately weighed (0.5 g) fresh plant leaf sample was homogenized in a tissue homogenizer with 10 ml of acetone as the extraction solvent. The homogenized sample was centrifuged at 12,000 rpm at 4 °C for 15 min. One ml of the separated supernatant was mixed with 4 ml of the acetone solvent. The solution mixture was analyzed for Chl-a, Chl-b, total chlorophyll, and carotenoid contents by spectrophotometry using the following Equations37:
A = Absorbance, Chl-a = Chlorophyll a, Chl-b = Chlorophyll b, Cx + c = Carotenoids.
Determination of antioxidant enzyme activity
Fresh root and leaf tissues (0.5 g) were ground into fine powder under liquid nitrogen and then mixed with 10 mL of pre-cooled phosphate buffer (50 mM, pH 7.8) containing 1.0% (w/v) PVP. The mixture was centrifuged at 8000 × g and 4 °C for 40 min. The obtained supernatant was used for the enzyme assay as crude enzyme preparation. Catalase (CAT)38, ascorbate peroxidase (APX)39, and peroxidase (POD)40 activities were assayed according to Ekinci et al., Nakano and Asada, and Chance and Machly, respectively. Since the addition of H2O2, changes in absorbance were monitored for 120 s at 240, 290, and 470 nm to measure CAT, APX, and POD activities, respectively.
Protein extraction
Protein and enzyme extracts were prepared following the previous method41. In brief, fresh leaves (0.5 g) were ground to a fine powder in liquid nitrogen using a pre-chilled mortar and pestle and then extracted in 3 mL of 0.2 M potassium phosphate buffer (pH 7.0), containing 0.1 mM of ethylenediaminetetraacetic acid (EDTA). The extract was centrifuged at 13,000 rpm and 4 °C for 20 min, and the supernatant was used for the protein activity assay. The total protein content for all samples was determined by the method of Bradford (1976) using bovine serum albumin as a standard42.
RNA extraction and expression patterns of nitrate-transporter genes
To prepare leaf RNA, leaves and root samples were collected separately from barley seedlings under ND stress (on 7 and 14 days) and normal conditions. Total RNA was extracted from nitrogen-stressed and normal leaves and roots using an RNX-Plus kit (Sinaclone) according to the manufacturer's instructions. The purity and concentration of RNA were determined by NanoDrop, and its quality was confirmed using the 1% agarose gel analysis. Then, cDNA was synthesized according to the instructions of a cDNA synthesis kit. Each gene was analyzed with three repetitions, where the actin gene was used as a reference gene. All primers used in the gene expression analysis were designed using the Oligo program (Table 3). Gene expression was examined with a real-time instrument using Cybergreen as described in the manufacturer's instructions. After normalization, the relative expression of genes was evaluated through 2-∆∆CT and the value of Ct for nitrate transporter genes was determined using actin as a reference gene. The qRT-PCR analysis was performed to determine the expression profiles of HvNRT2.1, HvNRT2.2, HvNRT2.3, HvNRT2.4, HvNRT2.5, HvNRT2.6, HvNRT2.8, HvNRT2.9, and HvNRT2.10 genes using leaf and root tissues under normal and ND treatments. The expression levels of these genes were also investigated at the seedling stage.
Statistical analysis
The TB tools were utilized to draw the heat map, which was used to display the differential expression of genes and the correlation between the physiological traits and gene expression. Statistical analyses were performed using SPSS version 20.0 statistical software. Significant variations between means were compared at P < 0.05 (Duncan's test). Statistical graphs were generated using GraphPad Prism version 9 software at a statistical significance of p < 0.05.
Data availability
All data generated or analyzed during this study are included in this present article.
References
Xu, X. et al. Nitrogen–potassium balance improves leaf photosynthetic capacity by regulating leaf nitrogen allocation in apple. Hortic. Res. 11, 253 (2024).
Näsholm, T., Kielland, K. & Ganeteg, U. Uptake of organic nitrogen by plants. New Phytol. 182, 31–48 (2009).
Abrol, Y. P., Chatterjee, S. R., Kumar, P. A. & Jain, V. Improvement in nitrogen use efficiency: Physiological and molecular approaches. Curr. Sci. 76, 1357–1364 (1999).
Rivero-Marcos M, Silva Jr GB, Ariz I (2023)Structural role of silicon-mediated cell wall stability for ammonium toxicity alleviation. In: Benefits of Silicon in the Nutrition of Plants. Cham: Springer International Publishing. 209–236.
Huang, Z. A. et al. Effects of nitrogen deficiency on gas exchange, chlorophyll fluorescence, and antioxidant enzymes in leaves of rice plants. Photosynthetica. 42, 357–364 (2004).
Agarwal, S., Sairam, R. K., Srivastava, G. C. & Meena, R. C. Changes in antioxidant enzymes activity and oxidative stress by abscisic acid and salicylic acid in wheat genotypes. Plant Biol. 49, 541–550 (2005).
Sivasankar, A. et al. Differential response of two wheat genotypes to nitrogen supply I. Ontogenic changes in laminae growth and photosynthesis. J. Agron. Crop Sci. 181, 21–27 (1998).
Binder, D. L., Sander, D. H. & Walters, D. T. Maize response to time of nitrogen application as affected by level of nitrogen deficiency. J. Agron. 92, 1228–1236 (2000).
Fowler, D. et al. The global nitrogen cycle in the twenty-first century. Philo. Trans. R Soc. Lond. B Biol. Sci. 368, 20130164 (2013).
Bi, Y. M. et al. Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant Cell Environ. 32, 1749–1760 (2009).
Dai, Z. C. et al. Strong invasive mechanism of Wedelia trilobata via growth and physiological traits under nitrogen stress condition. Plants 13, 355 (2024).
Filleur, S. et al. An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489, 220–224 (2001).
Kiba, T. et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell. 24, 245–258 (2012).
Li, W. et al. Dissection of the AtNRT2.1: AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433 (2007).
Lezhneva, L. et al. The Arabidopsis nitrate transporter NRT 2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80, 230–241 (2014).
Fan, X. et al. Plant nitrate transporters: From gene function to application. J. Exp. Bot. 68, 2463–2475 (2017).
Tong, Y., Zhou, J. J., Li, Z. & Miller, A. J. A two-component high-affinity nitrate uptake system in barley. Plant J. 41, 442–450 (2005).
Olšovská, K., Sytar, O. & Kováčik, P. Optimizing nitrogen application for enhanced barley resilience: A comprehensive study on drought stress and nitrogen supply for sustainable agriculture. Sustainability 16, 2016 (2024).
Ganie, A. H. et al. Nitrogen-regulated changes in total amino acid profile of maize genotypes having contrasting response to nitrogen deficit. Protoplasma 254, 2143–2153 (2017).
Forde, B. G. et al. Nitrogen signalling pathways shaping root system architecture: An update. Curr. Opin. Plant Biol. 21, 26 (2014).
Giehl, R. F., Gruber, B. D. & von Wirén, N. It’s time to make changes: Modulation of root system architecture by nutrient signals. J. Exp. Bot. 65, 778 (2014).
Zhao, L. et al. Physiological and transcriptional characteristics of banana seedlings in response to nitrogen deficiency stress. Horticulture 10, 290 (2024).
Rosado, S. I. P. et al. Nitrate/ammonium ratios and nitrogen deficiency impact on nutrient absorption and photosynthetic efficiency of Cedrela odorata. Nitrogen 5, 1–15 (2024).
Zhang, Y. et al. Estimation of vegetation photosynthetic capacity from space-based measurements of chlorophyll fluorescence for terrestrial biosphere models. Glob. Change Biol. 20, 3727–3742 (2014).
Cartelat, A. et al. Optically assessed contents of leaf polyphenolics and chlorophyll as indicators of nitrogen deficiency in wheat (Triticum aestivum L.). Field Crops Res. 91, 35–49 (2005).
Peng, S. et al. Increased N use efficiency using a chlorophyll meter on high-yielding irrigated rice. Field Crops Res. 47, 243–252 (1996).
Okamoto, M. & Okada, K. Differential responses of growth and nitrogen uptake to organic nitrogen in four gramineous crops. J. Exp. Bot. 55, 1577–1585 (2004).
Machado, J. et al. Enzymatic and non-enzymatic antioxidant responses of young tomato plants (cv Micro-Tom) to single and combined mild nitrogen and water deficit: not the sum of the parts. Antioxidants 12, 375 (2023).
Jacquot, A. et al. NRT2.1 C-terminus phosphorylation prevents root high affinity nitrate uptake activity in Arabidopsis thaliana. New Phytol. 228, 1038–1054 (2020).
Rivero-Marcos, M. et al. Plant ammonium sensitivity is associated with the external pH adaptation, repertoire of nitrogen transporters, and nitrogen requirement. J. Exp. Bot. 11, 106 (2024).
Zhang, X. et al. Functional characterization of the GhNRT2.1e gene reveals its significant role in improving nitrogen use efficiency in Gossypium hirsutum. Peer J. 11, 15152 (2023).
Cun, Z. et al. Identification of candidate genes and residues for improving nitrogen use efficiency in the N-sensitive medicinal plant Panax notoginseng. BMC Plant Biol. 24, 105 (2024).
Zhuo M, Sakuraba Y, Yanagisawa S (2024) Dof1. 7 and NIGT1 transcription factors mediate multilayered transcriptional regulation for different expression patterns of nitrate transporter2 genes under nitrogen deficiency stress. New Phytol. 24.
Hamaoka, N. et al. Genetic variations in dry matter production, nitrogen uptake, and nitrogen use efficiency in the AA genome Oryza species grown under different nitrogen conditions. Plant Prod. Sci. 16, 107–116 (2013).
Han, L., Sun, K., Jin, J. & Xing, B. Some concepts of soil organic carbon characteristics and mineral interaction from a review of literature. Soil Biol. Biochem. 94, 107–121 (2016).
Sumanta, N., Haque, C. I., Nishika, J. & Suprakash, R. Spectrophotometric analysis of chlorophylls and carotenoids from commonly grown fern species by using various extracting solvents. Res. J. Chem. Sci. 2231, 606X (2014).
Ekinci, M. et al. Determination of physiological indices and some antioxidant enzymes of chard exposed to nitric oxide under drought stress. Russ. J. Plant Physiol. 67, 740–749 (2020).
Nakano, Y. & Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. PCP 22, 867–880 (1981).
Chance, B. & Maehly, A. C. Assay of catalases and peroxidases. Methods Enzymol. 2, 764–775 (1955).
Bradford, M. Bradford method. The protein estimation kit from Boston Bio Products is based on the method of Bradford. Anal. Biochem. 72, 248–254 (1976).
Chang, C. J. & Kao, C. H. H2O2 metabolism during senescence of rice leaves: changes in enzyme activities in light and darkness. Plant Growth Regul. 25, 11–15 (1998).
Acknowledgements
The authors are thankful to the Department of Cell & Molecular Biology, Faculty of Life Sciences and Biotechnology, Shahid Beheshti University,Tehran, Iran and Elite National Foundation for receiving necessary help for this project work.
Author information
Authors and Affiliations
Contributions
A.S. and Z.H. designed this work. Z.H. performed the experiments. Z.H. and A.S. carried out data analysis. All authors discussed the results. Z.H., H.G.H. and A.S. wrote the manuscript. A.S. edited the final version of the manuscript. Correspondence should be addressed to A.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hajibarat, Z., Saidi, A., Ghazvini, H. et al. Investigation of morpho-physiolgical traits and gene expression in barley under nitrogen deficiency. Sci Rep 14, 8875 (2024). https://doi.org/10.1038/s41598-024-59714-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-59714-z
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.