Abstract
Glioblastoma (GBM) is an aggressive primary CNS malignancy and clinical outcomes have remained stagnant despite introduction of new treatments. Understanding the tumor microenvironment (TME) in which tumor associated macrophages (TAMs) interact with T cells has been of great interest. Although previous studies examining TAMs in GBM have shown that certain TAMs are associated with specific clinical and/or pathologic features, these studies used an outdated M1/M2 paradigm of macrophage polarization and failed to include the continuum of TAM states in GBM. Perhaps most significantly, the interactions of TAMs with T cells have yet to be fully explored. Our study uses single-cell RNA sequencing data from adult IDH-wildtype GBM, with the primary aim of deciphering the cellular interactions of the 7 TAM subtypes with T cells in the GBM TME. Furthermore, the interactions discovered herein are compared to IDH-mutant astrocytoma, allowing for focus on the cellular ecosystem unique to GBM. The resulting ligand-receptor interactions, signaling sources, and global communication patterns discovered provide a framework for future studies to explore methods of leveraging the immune system for treating GBM.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is one of the most aggressive primary CNS malignancies. Despite advances in treatment protocols, clinical outcomes have not followed, as evidenced by a 5-year survival rate near 5%2. Key obstacles remain, including an understanding of the heterogeneous tumor microenvironment (TME). Recent focus has shifted towards the cellular relations in this system, mainly the surrounding stromal cells of which macrophages are the most prominent and significant in disease progression1,3,4.
Macrophages and microglia (local CNS macrophages derived from the yolk sac) are both important components of the immune response. While microglia have self-renewal capacity and maintain brain homeostasis with minimal immune stimulation5,6, bone-marrow-derived macrophages infiltrate the brain only in pathological conditions7. Indeed, in GBM, the monocyte-derived macrophages may compose as much as 50% of the total mass8. However, unlike normal macrophages, these tumor-associated macrophages (TAMs) behave differently, altering the bridge between innate and adaptive immune responses and causing states of pathologic TAM-mediated immunosuppression.
A major component of the TAM-mediated immunosuppression involves the lymphoid lineage, namely T cells. It has been observed that TAMs may impede T cell functionality, promote T cell exhaustion, and inhibit T cell migration9. As macrophages exhibit high degrees of plasticity, they have historically been viewed in the M1 (pro-inflammatory) or M2 (anti-inflammatory) phenotypes, with the M1 macrophages associated with anti-tumorigenic effects. It is now accepted that this dichotomous nomenclature oversimplifies TAM plasticity, especially in a dynamic setting such as the TME of GBM10,11,12. To truly appreciate the cellular interactions of TAMs and T cells in GBM, a framework that includes a continuum of TAM activation states must be used.
Recently, 7 TAM subtypes were found to be preserved in almost all cancer types and were characterized using single-cell multi-omics technologies. These 7 TAM subsets, based on signature genes and enriched pathways, have been categorized as follows: interferon-primed TAMs (IFN-TAMs), immune regulatory TAMs (reg-TAMs), inflammatory cytokine-enriched TAMs (inflam-TAMs), lipid-associated TAMs (LA-TAMs), pro-angiogenic TAMs (angio-TAMs), resident-tissue macrophage-like TAMs (RTM-TAMs), and proliferating TAMs (prolif-TAMs)12.
Prior research investigating TAMs in GBM have identified specific types that are correlated with clinical and pathological characteristics1. However, these investigations have relied on the antiquated M1/M2 paradigm of macrophage polarization and have neglected to account for the continuum of TAM states in GBM, as well as their interaction with T cells. Hence, the current study leveraged single-cell RNA sequencing data obtained from adult IDH-wildtype GBM in order to analyze the cellular interactions between the seven TAM subtypes and T cells within the GBM TME. Furthermore, comparing the interactions to those of IDH-mutated astrocytomas emphasizes the signaling ecology specific to GBM. The identified ligand-receptor interactions, signaling sources, and global communication patterns establish a foundation for subsequent research endeavors that aim to investigate strategies for harnessing the immune system in the treatment of GBM.
Methods
Data acquisition
Single-cell RNA sequencing data matrices and metadata comprising myeloid and T cells from 7 newly diagnosed IDH wildtype GBM samples and 1 IDH mutant astrocytoma sample derived from a previous study were used for the present analysis (Gene Expression Omnibus accession GSE182109)13. Another dataset including immune cells from GBM samples was examined separately for in silico validation (Gene Expression Omnibus accession GSE163120)14. Additionally, previously published GBM spatial transcriptomics data (sample 269_T, available at https://doi.org/10.5061/dryad.h70rxwdmj) was computationally examined to explore the spatial distribution of gene expression of certain cell type patterns in the tissue extracted from these samples with respect to the barcode-spot’s spatial dimensions15. The ‘AUCell’ R package was used to annotate the TAM subpopulations from the original datasets. Gene signatures derived from previous literature12 (Table 1) was used for TAM subpopulation classification. The R packages ‘celldex’ and ‘SingleR’ were used to classify the other immune cell populations, including T cells, into further subtypes using the ‘MonacoImmuneData’ reference index, which contains normalized expression values of bulk RNA-seq samples of 29 sorted immune cell populations, allowing for high resolution classification of immune cell transcriptomes16.
Ligand-receptor analysis
Cell communication analysis was based on CellChat17. The CellChatDB was used as the reference database of literature-supported ligand-receptor interactions, containing more than 1900 validated and manually curated interactions including paracrine and autocrine secreted signaling interactions. Using this database of known ligand-receptor interactions, CellChat initially identifies differentially overexpressed receptors and ligands in each provided cell type. Each interaction is subsequently associated with a probability value, modelled by the law of mass action based on the average expression values of a receptor of one receiver cell type and the average expression values of the ligand of the counterpart cell type.
CellChat was run using ‘trimean’, a robust statistical method for calculating the average gene expression per cell type. This method assembles fewer interactions but performs better for predicting stronger interactions which is advantageous when selecting those interactions for further experimental validations. Lastly, the significance of these interactions is identified by randomly permuting the cell type labels and subsequently recalculating the interaction probability17. Comparisons between IDH wildtype and mutant for identifying up-regulated and down-regulated signaling was performed by comparing the communication probability between two datasets for each ligand-receptor pair and each pair of cell groups. Interactions with p value < 0.05 were considered significant.
Identifying major signaling sources and global communication patterns
To examine dominant senders and receivers within the interactome, the present study leveraged metrics derived from graph theory, which were previously utilized for social network analysis17,18. Specifically, weighted-directed networks and associated metrics, including out-degree and in-degree, were used to discover dominant intercellular interaction senders and receivers. In a weighted-directed network, the weights (i.e., strengths of interactions) are defined as the calculated communication probabilities based on the gene expression of ligands and receptors. Out-degree is calculated as the sum of these communication probabilities of outgoing signaling from an individual cell group. In-degree is computed as the sum of communication probabilities of incoming signaling to a cell group. These scores were used to identify the dominant cell senders and receivers, respectively, of intercellular signaling networks in the weighted-directed network17. Additionally, information flow was calculated for a signaling pathway by summing the communication probabilities amongst all pairs of cell groups in the predicted network17,19.
To identify communication patterns among all signaling pathways, non-negative matrix factorization was carried out, with the number of patterns based on the Cophenetic and Silhouette metrics, which measure the stability for a particular number of patterns via hierarchical clustering of the derived consensus matrix17,20. Plainly, outgoing communication patterns show how sender cell types coordinate with each other and how they work with certain signaling pathways to support communication. Correspondingly, incoming patterns reveal how target cell types (i.e., cells receiving signals) coordinate with each other to respond to incoming signaling.
Results
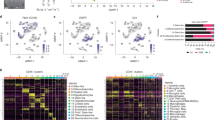
Using dimensionality reduction, the TAMs from GBM and IDH-mutant glioma showed distinct clustering profiles as well as areas of overlap (Fig. 1A). All seven types of TAMs were found in both types of gliomas (Fig. 1B). IDH-wildtype GBM harbored more LA-TAMs, inflammatory TAMs, proliferative TAMs, but had less RTM microglia-like TAMs. Of the 46,550 total cells sequenced from the original datasets, 3957 T cells were identified (8.5%) and subsequently used for further interaction analyses (Supplementary Figure 1).
Predicted ligand-receptor interactions between T lymphocytes and TAMs
To predict significant ligand-receptor interactions between TAMs and T cells, a comprehensive dataset including single-cell gene expression data from both IDH wildtype primary GBM and IDH-mutant gliomas was used13. To understand which cell types are dominant senders (i.e., cell types sending signals) and which are dominant receivers (i.e., cell types receiving signals) with respect to each ligand-receptor interaction analyzed, incoming and outgoing interactions strengths (weights) were calculated for both GBM and glioma (Fig. 2A). In both IDH wildtype and mutant, angio-TAMs displayed robust incoming and outgoing signaling strength. In the IDH-wildtype, all seven TAM types displayed strong outgoing interaction strength compared to IDH-mutant, where only inflammatory TAMs and LA-TAMs showed strong outgoing interaction strength.
(A) Scatterplot displaying dominant senders and receiver cell type. The total outgoing and incoming communication probability associated with each cell group is displayed on x-axis or y-axis, respectively. Count is displayed by dot size which is proportional to the number of predicted signaling links associated with each cell group. (B) Stacked bar plot and unstacked bar plot (C) showing conserved and context specific signaling pathways by comparing information flow for each signaling pathway. (D) Scatterplot showing specific signaling changes of TAM populations. Positive values indicate increase in IDH wildtype GBM compared to IDH mutant astrocytoma.
Identifying signaling pathways specific to GBM
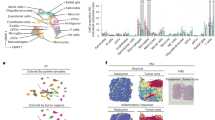
Next, we then sought to identify those signaling pathways that were conserved and those that were specific to GBM by calculating the information flow within signaling pathways, defined as the sum of communication probabilities amongst the total pairs of cell groups in the network (i.e., the amount of activity occurring in the pathway) (Fig. 2B). Compared to the IDH-mutant astrocytoma, GBM exhibited increased information flow in multiple pathways including GALECTIN, COMPLEMENT, MIF, SPP, and PTN. When considering the absolute information flow in each pathway (Fig. 2C), greater information flow was observed in CCL, ANNEXIN, MIF, GALECTIN, COMPLEMENT, and SPP1 pathways. These flow states imply that although IDH-wildtype GBM has increased activity in these pathways relative to IDH-mutant astrocytoma, both types of gliomas have substantial activity occurring within these signaling pathways.
Since IDH-wildtype GBM displayed prominent activity of TAMs (Fig. 2A) and displayed specificity for certain pathways (Fig. 2B, C), we then sought to identify the specific signaling changes of the different TAM populations between GBM and IDH-mutant (Fig. 2D). Angio-TAMs and LA-TAMs were observed to have shared signaling amongst both glioma types in SPP1 and MIF pathways. However, the other five TAM subtypes showed increased signaling in SPP1 specific to GBM (Fig. 2D). Overall, the seven TAM populations were observed to be contributing in an increased capacity to pathways such as SPP1, COMPLEMENT, GALECTIN and PTN compared to their counterparts in IDH-mutant astrocytoma.
Further examination of each pathway yielded numerous significant ligand-receptor interactions (Fig. 3A, B). All types of TAMs exhibited MIF and SPP1 binding interactions with ligands on T cells such as CD44 and ITGA4 + ITGB1 (Fig. 3A). Conversely, T cell ligands interacting with receptors on TAMs commonly included MIF-(CD74 + CD44) and ANXA1-FPR1 (Fig. 3A). To further verify these interactions, another dataset that included was analyzed independently in a similar fashion14. The ligand-receptor interactions were corroborated by the second dataset, as interactions including SPP1, MIF, LGALS9 between TAMs to T cells were found to be significant (Fig. 4A, B). When deciphering the ligand-receptor interactions in the context of cell-to-cell contact (Fig. 5), numerous significant interactions between HLA class I molecules and CD8 were observed in GBM. Such HLA class I molecules were indicated to be on TAMs, including HLA-A, B, C, E, and F. Additional interactions from TAMs to effector CD8 T cells such as CD99-CD99 were also seen. Activated leukocyte cell adhesion molecule (ALCAM) interacting with CD6 on T cells were witnessed to be significant in most of the interactions from TAMs to T cells (Fig. 5).
(A) Chord diagram visualizing cell–cell communication for significant ligand-receptor interactions from TAM populations (senders, bottom arc) to T cell populations (receivers, top arc). (B) Chord diagram visualizing significant ligand-receptor interactions from T cell populations (senders, bottom arc) to TAM populations (receivers, top arc).
Bubble plot showing all the significant secreted signaling interactions (L-R pairs) (y-axis) from TAM populations (senders) to T cell populations (receivers) (x-axis). The dot color and size represent the calculated communication probability (represents interaction strength) and p-values (represents statistical significance). Non-significant interactions represented by blank spaces. (A) Abdelfattah et al., 2022 dataset. (B) Additional independent analysis using Pombo Antunes et al., 2021 dataset.
Same as Fig. 4 but for cell-to-cell contact interactions.
Global communication patterns of TAMs
Along with understanding ligand-receptor interactions in the tumor microenvironment, it is crucial to analyze how different cell types coordinate with each other in terms of signaling. To address this, a pattern recognition methodology was employed to identify global communication patterns of TAMs and T cells (see Methods). These communication patterns can be thought of as assigning cell types to signaling pathways for either outgoing signaling (considering cells as senders) or incoming signaling (considering cells as receivers) (Fig. 6A, B). The outgoing cell signaling patterns were broadly identified into 3 distinct categories. Most TAMs contribute to pattern 1, while pattern 2 was mainly contributed by angio-TAMs and T regulatory cells (Fig. 6A). Pattern 1 was mainly composed by pathways involving TNF, GALECTIN, SPP1, and COMPLEMENT while pattern 2 mainly composed of RESISTIN and ANGPTL signaling (Fig. 6A). For the incoming signaling pathways in IDH wildtype GBM, 3 discernable patterns also emerged (Fig. 6B). Interestingly, angio-TAMs displayed pattern 3, as opposed to the other TAMs which displayed a pattern 1 incoming signaling. Pattern 3 is mostly contributed by VISFATIN, IL-1, ANGPTL and CD40.
Global communications analysis for (A) outgoing and (B) incoming communication patterns. Cell patterns heatmap display which cell types contribute to which type of pattern. Communication patterns heatmap displays which signaling pathways contribute to the pattern. The dot plots show how each cell group contributed to the signaling pathways, where the dot size is proportional to the contribution score to show association between cell group and their enriched signaling pathways. A higher contribution score implies the signaling pathway is more enriched in the respective cell type.
These communication results imply that TAMs in GBM may depend on numerous signaling networks that may overlap in function while certain TAMs, such as angio-TAMs, rely on fewer and more heterogenous communication patterns. Additionally, leveraging these incoming and outgoing patterns may provide insight into autocrine vs. paracrine pathways for a given cell type in the GBM cellular landscape.
T cell exhaustion signature and spatial transcriptomics
Given this interplay amongst TAMs and T cells, especially within the context of T cell exhaustion in GBM21, a better understanding of the spatial interactions of these cell types were explored. First, examining genes related to T cell exhaustion revealed an increased expression in T cells within GBM (Fig. 7). Specifically, T regulatory lymphocytes in GBM exhibited increased expression of ICOS, CTLA4, TIGIT, IL2RA, and IL10RA (Fig. 7). To evaluate further, spatial transcriptomic data from a GBM dataset15 was examined by transposing the gene signatures of T cell exhaustion and the TAMs analyzed in the present study (Fig. 8). The T cell signature used included persistent expression of multiple inhibitory receptors, such as PD-1, LAG-3, TIM-3, CTLA-4, and TIGIT21. The spatial plots displayed a prominent overlap amongst TAM signatures and the T cell exhaustion gene signature, with a higher density and expression of T cell exhaustion related genes in spaces occupied by TAM signatures (Fig. 8).
Discussion
IDH mutant gliomas and GBM have distinct immune profiles with a diverse repertoire of TAMs
All seven subsets of TAMs were found in both IDH wildtype GBM and mutant gliomas. Angio-TAMs have an angiogenic signature associated with significant immune suppression and tumor cell proliferation. Though found in proportionally smaller numbers, they were observed to be the dominant receivers and senders in the population communicating with the lymphocyte population. Angio-TAMs were rarely encountered in IDH mutant gliomas. Both IDH mutant and IDH WT gliomas have increased representation of myeloid cells with lymphocyte population increased in the IDH WT tumors4. RTM-TAMs formed the largest component of TAMs in the IDH mutant variety. RTM-TAMs are very similar to normal RTMs and a putative role for them in tumor invasiveness in lung cancer has been suggested22. LA-TAMs and proliferative TAMs were also seen in larger numbers in the WT high grade gliomas. LA-TAMs express more lipid related and oxidative phosphorylation genes which include APOC1, APOE, ACP5 and FABP5.Lipid catabolism in macrophages is associated with IL-4 associated pathways and anti-inflammatory phenotype23. Inflammatory TAMs have an inflammatory signature and express inflammatory cytokines including IL1B, CXCL1/2/3/8, CCL3, and CCL3L1. Inflammatory TAMs are known to play an active role in the recruitment of immune cells, including granulocytes, into the TME. Regulatory TAMs, which were found more often in GBM, seemed to have a profile consistent with microglia and appeared to play a more quiescent role in activating lymphocytes. This was corroborated by Venteicher et al. who demonstrated microglia in IDH mutant glioma TME were associated with a less malignant phenotype24.
Effector memory CD8 T cells were more often encountered in WT glioma while NK cells were more common in IDH mutant glioma. Th1, Th2 and Th 17 CD4+ T cells were hardly seen in IDH mutant gliomas, compared to IDH-WT tumors. Vd2 gd T cells were present abundantly in both groups.
TAMs are associated with T cell and NK cell exhaustion in the TME
T lymphocyte exhaustion is a well-known phenomenon in GBM with cells expressing several inhibitory checkpoint receptors, including PD-1, LAG-3 and CTLA-425. Leveraging spatial transcriptomics revealed that the signatures of all seven TAM subtypes were associated with a strong expression of genes related to T cell exhaustion. Ravi et al. previously identified a subset of TAMs that secreted IL-10 and HMOX1+ and contributed to T cell exhaustion in glioblastoma26. These cells were characterized by the expression of CD163, CCL4, APOE and HLA–DRE. In our dataset, they would map to angio-TAMs, LA-TAMS and Inflam-TAMs. The TAMs were observed to produce an immunosuppressive environment by secreting ligands such as MIF, Galectin 9 and SPP1 and affecting cell-to-cell contact and are described in subsequent sections.
CD86, HAVCR2 (TIM3), TGFB1 were significantly expressed in TAMs. TIGIT was upregulated in central memory CD8 T cells. ICOS (CD278), CTLA4 and TIGIT were robustly expressed in T regs. IL-10 RA, which codes for the IL-10 receptor, is well-known to induce immunosuppression. It was strongly expressed in all the immune cells consistent with prior studies showing that IL-10 contributes to the immunosuppressed environment in gliomas26.
Transforming growth factor-beta (TGF-β), which promotes an immunosuppressive milieu, was also seen to be expressed by all TAMs in secreted signaling interactions (Fig. 7). PRDM1, known to regulate PD-L1 levels, resulting in CD8 + T cell exhaustion, was also expressed in all immune cells examined27. CD96, an immune checkpoint ligand, was strongly expressed in T and NK lymphocytes within GBM samples. VSIR, an important checkpoint modulator in AML, was highly expressed in both IDH mutant and GBM TAM populations28,29. FGD4, albeit not well studied, was also well represented in the same group of cells30,31 (Fig. 7). Furthermore, TAMs interact with various immune cells within the tumor microenvironment through other mechanisms, which include expression of Indoleamine 2,3-dioxygenase (IDO) and the production of Arg1, which is beyond the scope of the current analysis.
TAMs in GBM secrete a common group of ligands which are immunomodulatory
Immune cells in the TME of GBM secrete a variety of immunomodulatory molecules21,26. During the present analysis, certain prominent signaling pathways common amongst TAMs and T cells in GBM were revealed, including galectin, complement, MIF, SPP, and PTN signaling pathways (Fig. 2–4). In addition, when considering the absolute information flow in each pathway, CCL and ANNEXIN also assumed importance (Fig. 2). All these signaling pathways account for significant immune suppression and support tumor proliferation.
Regardless of the TAM subtype, SPP1 (Osteopontin) seemed to be very significant in outgoing and incoming signaling (Fig. 2B–D). SPP1 suppresses T cell function through the IRF8-OPN-CD 44 axis32. MIF (Macrophage Inhibitory Factor) also can act to reduce CD8 + cytotoxic lymphocyte (CTL) activity33,34. We found that there was significant upregulation of the MIF ligand—CD74/CD44 pair, which can recruit macrophages and foster an immunosuppressive niche. Blocking this interaction in TAMs and dendritic cells in melanoma showed decreased immunosuppression35. MIF also interacts with immune checkpoint molecules like PD-L1, contributing to T cell exhaustion and immune evasion while suppressing cytotoxic T cells and NK cells33. Complement secreted by TAMs is increasingly seen as immunosuppressive and promotes tumor growth. This immunosuppressive effect is mediated through multiple pathways, which include the increased expression of molecules such as PDL-1, IL-10, Arg-1, and TGF-β136.
Our results also revealed significant expression of interactions involving galectins and the galectin signaling pathways (Figs. 2–4, 6). Galectin 1 is highly expressed in glioblastoma stem cells and is important for immunomodulation and angiogenesis31,32,33. Galectin 9 is associated with T cell exhaustion in the tumor environment38,39. The binding of TIM3 to its ligand, galectin-9 induces T cell apoptosis33. Galectin binds to the CD45 receptor, as shown in the ligand interaction, and CTLA 4 to reduce T cell proliferation and induce T cell apoptosis43. Galectin is also involved in the formation and stability of regulatory T cells (suppressors of antitumor immunity) through the CD44 receptor in association with TGF-β. The CD44 receptor is crucial in mediating the action of SPP1, Galectin and MIF. CD44 is glycoprotein which is used as a marker for cancer stem cells (CSC) and has significant role in the maintenance and progression of solid tumors40. CD44 is also involved in the positive regulation of the immune checkpoint molecule PD-L1, macrophage infiltration of tumors and contributes to the antitumor activity of TAMs41.
The annexin pathway is also revealed to be crucial in the secreted signaling interactions between TAMs and T cells (Figs. 2–4, 6). Annexin 1 is expressed in glioblastoma and known to promote tumor growth while fostering an immunosuppressive environment42,43. Treatment of macrophages with AnxA1 results in decreased iNOS expression, increased IL-10 levels and decreased IL-12 mRNA levels and an anti-inflammatory phenotype42. CCL (chemokine ligands) secreted can also produce an environment that is favorable to tumor progression or result in anti-tumor activity. The recruitment of monocytes from peripheral blood and differentiation into TAMs are dependent on the CCR5-CCL5 and CCR2-CCL2 chemokine axes. TAMs also secrete CCL17, CCL22, and CCL18 which attract regulatory T cells44. Pleiotrophin (PTN), another significant result in our analyses, is known to be secreted by TAMs and enhance glioblastoma cell proliferation through its receptor PTPRZ1 expressed on glioma stem cells (GSCs)45. PTN also contributes to angiogenesis in gliomas and increased expression of PTN is associated with poor survival in glioma patients46.
Visfatin/NAMPT, originally isolated from peripheral lymphocytes, can induce monocytes to transform into TAMs with CD163 expression. Indeed, angio-TAMs express CD163 strongly on their surface (Table 1)12. These TAMs also secrete the chemokine CXCL1 which are known to increase invasion and migration of cancer cells along with angiopoietins that increase angiogenesis and promote tumor growth in glioblastomas47,48 .
Angiopoietin-like proteins (ANGPTLs) are structurally like angiopoietins. We found multiple interactions involving these molecules in the current analysis. Angiopoietin-like 2 (ANGPTL2) fosters the conversion of TAMs into a less inflammatory subset in NSLC. It also correlates positively with TAM infiltration and a poor prognosis47,49. Another family member, angiopoietin-like 4 (ANGPTL4) blunts the polarization of macrophages toward the proinflammatory phenotype and decreases immune surveillance in tumor progression by downregulating CD8 T cell activation50. Resistin was another key mediator in our analysis. Mostly expressed by macrophages in humans, it has potent pro-inflammatory properties and is shown to promote cancer progression while inducing expansion of regulatory T cells, which have an immunosuppressive role51,52.
TAMs in WT modulate T and NK lymphocytes through cell-to-cell contact
TAMs also communicate with lymphocytes through cell-to-cell contact (Fig. 5). Since the TAMs provide an immunosuppressive environment, it could be assumed that these interactions are inhibitory. Surprisingly the most prominent of these interactions center around the MHC Class I molecules, which reiterates recent insights into how TAMs regulate the immune environment53,54. Our results displayed that HLA—A, B, C and E interact with CD8 receptors significantly on central memory, effector memory, and terminal effector T cells. Indeed, HLA—A, B, C have been shown to negatively impact the function of NK cells and cytotoxic T lymphocytes through engagement with LILRs and lymphocytic KIR receptors, resulting in immunosuppression, anergy, and T cell exhaustion53,54. HLA-E on the surface of TAMs also bind to the inhibitory receptor CD94/NKG2A found on both NK cells and cytotoxic lymphocytes55. With regards to MHC class II molecules, HLA—DRB5 and CD4+ cells also interacted in cell-to-cell contact (Fig. 5). HLA-DRB5 has been found to be restrictive in presenting tumor antigens on CD 4+ cells56.
This interaction is of critical importance because it forms the frontline against cancer propagation. Unfortunately, in advanced cancers, such as GBM, the TAMs seem to be tumorigenic with regards to their immunosuppressive role in the tumor microenvironment and T cell activation. While the roles of Leukocyte Immunoglobulin-Like Receptors (LILRs) and Killer cell Immunoglobulin-like Receptors (KIRs) were initially evolved to quell an overactive immune response, tumors have hijacked the response for immune evasion and T cell tolerance, allowing the tumor cells to grow unabated.
Other interactions, such as those involving CLEC2D and the receptor KLRB1 were also seen. Cytotoxic T cells in glioblastoma express the NK lymphocytic gene KLRB1. TAMs express the ligand CLEC2D on the surface which serves as an inhibitor for the KLRB1 receptor to suppress their tumor lytic function57. The homophilic CD99-CD99 interaction was demonstrated by most TAMs in our analyses. Previous studies show CD99 ligation presents an apoptotic feature in developing T lymphocytes, with increased CD99 expression associated with immune adaptation dominated by TAMs in gliomas58. The TAMs also interacted with lymphocytes through expression of CD86, which acts as the dominant ligand for CD4 + FoxP3 + regulatory T cell survival and proliferation, interacting with CD28 and CTLA4 receptors on the regulatory T cells59. It is also crucial to consider the paucity of T lymphocytes in GBM, which have been colloquially termed ‘immunologically cold.’ However, the present study exemplifies that those T lymphocytes and myeloid cells that exist within GBM are indeed involved in crucial immunomodulatory interactions.
Conclusion
The immune landscape in glioblastoma is complex and, most importantly, not static. It evolves along with the progression of the tumor. Immunosuppressive macrophages dominate the later stages of glioma evolution. We have identified several subsets of macrophages, among which angiogenic TAMs play a very significant role. The immunosuppressive molecules galectin, SPP1, MIF, and complement impede the T cell response. In addition to their immunosuppressive role in the TME, the present study supports the view that TAMs directly facilitate glioma cell proliferation through their secreted ligands, such as galectin and SPP1, along with direct cell-to-cell contact mechanisms.
Immunotherapeutic agents, including checkpoint inhibitors and CAR T cell therapies, have not shown encouraging utility in GBM, which is consistent with the immunologically ‘cold’ nature of GBM, implying that the microenvironment is pervaded by immunosuppressive TAMs60,61. TAMs seem to be critical in the trifecta of reducing macrophage activation, inhibiting cytolytic function in NK and cytotoxic cells, and reverting T cells to immunosuppression. Targeting TAMs in cancer, perhaps through “re-education into a pro-inflammatory state, is critical to exploring if future immunotherapies should hope to have any substantial effect on outcomes.
Data availability
All data analyzed in the present study is available publicly in the repositories stated in the manuscript (see Methods): Gene Expression Omnibus accessions GSE182109, GSE163120, and https://doi.org/10.5061/dryad.h70rxwdmj.
References
Andersen, J. K., Miletic, H. & Hossain, J. A. Tumor-associated macrophages in gliomas: Basic insights and treatment opportunities. Cancers 14, 1319. https://doi.org/10.3390/cancers14051319 (2022).
Alexander, B. M. & Cloughesy, T. F. Adult glioblastoma. J Clin Oncol 35, 2402–2409. https://doi.org/10.1200/JCO.2017.73.0119 (2017).
Wei, J. et al. Immune biology of glioma-associated macrophages and microglia: Functional and therapeutic implications. Neuro-Oncology 22, 180–194. https://doi.org/10.1093/neuonc/noz212 (2020).
Klemm, F. et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643-1660.e17. https://doi.org/10.1016/j.cell.2020.05.007 (2020).
Wlodarczyk, A. et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36, 3292–3308. https://doi.org/10.15252/embj.201696056 (2017).
Hoeffel, G. & Ginhoux, F. Ontogeny of tissue-resident macrophages. Front. Immunol. 6, 486. https://doi.org/10.3389/fimmu.2015.00486 (2015).
Murray, P. J. & Wynn, T. A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737. https://doi.org/10.1038/nri3073 (2011).
Hambardzumyan, D., Gutmann, D. H. & Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 19, 20–27. https://doi.org/10.1038/nn.4185 (2016).
Woroniecka, K. I., Rhodin, K. E., Chongsathidkiet, P., Keith, K. A. & Fecci, P. E. T-cell dysfunction in glioblastoma: Applying a new framework. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 24, 3792–3802. https://doi.org/10.1158/1078-0432.CCR-18-0047 (2018).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288. https://doi.org/10.1016/j.immuni.2014.01.006 (2014).
Szulzewsky, F. et al. Human glioblastoma-associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia 64, 1416–1436. https://doi.org/10.1002/glia.23014 (2016).
Ma, R.-Y., Black, A. & Qian, B.-Z. Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol. 43, 546–563. https://doi.org/10.1016/j.it.2022.04.008 (2022).
Abdelfattah, N. et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 13, 767. https://doi.org/10.1038/s41467-022-28372-y (2022).
Pombo Antunes, A. R. et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 24, 595–610. https://doi.org/10.1038/s41593-020-00789-y (2021).
Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639-655.e13. https://doi.org/10.1016/j.ccell.2022.05.009 (2022).
Monaco, G. et al. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 26, 1627-1640.e7. https://doi.org/10.1016/j.celrep.2019.01.041 (2019).
Jin, S. et al. Inference and analysis of cell-cell communication using cell chat. Nat. Commun. 12, 1088. https://doi.org/10.1038/s41467-021-21246-9 (2021).
Cordeiro, M., Sarmento, R. P., Brazdil, P. & Gama, J. Evolving networks and social network analysis methods and techniques. In Social Media and Journalism—Trends, Connections, Implications (eds Višňovský, J. & Radošinská, J.) (InTech, 2018).
Landherr, A., Friedl, B. & Heidemann, J. A critical review of centrality measures in social networks. Bus. Inf. Syst. Eng. 2, 371–385. https://doi.org/10.1007/s12599-010-0127-3 (2010).
Gaujoux, R. & Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 11, 367. https://doi.org/10.1186/1471-2105-11-367 (2010).
Watowich, M. B., Gilbert, M. R. & Larion, M. T cell exhaustion in malignant gliomas. Trends Cancer 9, 270–292. https://doi.org/10.1016/j.trecan.2022.12.008 (2023).
Casanova-Acebes, M. et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature 595, 578–584. https://doi.org/10.1038/s41586-021-03651-8 (2021).
O’Neill, L. A. J., Kishton, R. J. & Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565. https://doi.org/10.1038/nri.2016.70 (2016).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478. https://doi.org/10.1126/science.aai8478 (2017).
Woroniecka, K. et al. T Cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 24, 4175–4186. https://doi.org/10.1158/1078-0432.CCR-17-1846 (2018).
Ravi, V. M. et al. T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat. Commun. 13, 925. https://doi.org/10.1038/s41467-022-28523-1 (2022).
Li, Q. et al. PRDM1/BLIMP1 induces cancer immune evasion by modulating the USP22-SPI1-PD-L1 axis in hepatocellular carcinoma cells. Nat. Commun. 13, 7677. https://doi.org/10.1038/s41467-022-35469-x (2022).
Yao, K., Zhou, E., Schaafsma, E., Zhang, B. & Cheng, C. Immune checkpoint gene VSIR predicts patient prognosis in acute myeloid Leukemia and myelodysplastic syndromes. Cancer Med. 12, 5590–5602. https://doi.org/10.1002/cam4.5409 (2023).
Liu, Y. et al. Identify the prognostic and immune profile of VSIR in the tumor microenvironment: A pan-cancer analysis. Front. Cell Dev. Biol. 10, 8478 (2022).
Shiravand, Y. et al. Immune checkpoint inhibitors in cancer therapy. Curr. Oncol. 29, 3044–3060. https://doi.org/10.3390/curroncol29050247 (2022).
Bossan, A. et al. Expression of FGD4 positively correlates with the aggressive phenotype of prostate cancer. BMC Cancer 18, 1257. https://doi.org/10.1186/s12885-018-5096-9 (2018).
Klement, J. D. et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J. Clin. Investig. 128, 5549–5560. https://doi.org/10.1172/JCI123360 (2023).
Yan, X., Orentas, R. J. & Johnson, B. D. Tumor-derived macrophage migration inhibitory factor (MIF) inhibits T lymphocyte activation. Cytokine 33, 188–198. https://doi.org/10.1016/j.cyto.2006.01.006 (2006).
Regulation of the CTL Response by Macrophage Migration Inhibitory Factor. J. Immunol. American Association of Immunologists. https://journals.aai.org/jimmunol/article/166/2/747/70562/Regulation-of-the-CTL-Response-by-Macrophage. Accessed 14 Aug 2023.
Roumenina, L. T. et al. Tumor cells hijack macrophage-produced complement C1q to promote tumor growth. Cancer Immunol. Res. 7, 1091–1105. https://doi.org/10.1158/2326-6066.CIR-18-0891 (2019).
Zhang, R., Liu, Q., Li, T., Liao, Q. & Zhao, Y. Role of the complement system in the tumor microenvironment. Cancer Cell Int. 19, 300. https://doi.org/10.1186/s12935-019-1027-3 (2019).
Ajarrag, S. & St-Pierre, Y. Galectins in glioma: Current roles in cancer progression and future directions for improving treatment. Cancers 13, 5533. https://doi.org/10.3390/cancers13215533 (2021).
Okoye, I. et al. Galectin-9 expression defines exhausted T cells and impaired cytotoxic NK cells in patients with virus-associated solid tumors. J. Immunother. Cancer 8, e001849. https://doi.org/10.1136/jitc-2020-001849 (2020).
Holderried, T. A. W. et al. Molecular and immune correlates of TIM-3 (HAVCR2) and galectin 9 (LGALS9) mRNA expression and DNA methylation in melanoma. Clin. Epigenet. 11, 161. https://doi.org/10.1186/s13148-019-0752-8 (2019).
Thapa, R. & Wilson, G. D. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem Cells Int. 2016, 2087204. https://doi.org/10.1155/2016/2087204 (2016).
Liu, S. et al. CD44 is a potential immunotherapeutic target and affects macrophage infiltration leading to poor prognosis. Sci. Rep. 13, 9657. https://doi.org/10.1038/s41598-023-33915-4 (2023).
Araújo, T. G. et al. Annexin A1 as a regulator of immune response in cancer. Cells 10, 2245. https://doi.org/10.3390/cells10092245 (2021).
Yang, Y. et al. Annexin 1 released by necrotic human glioblastoma cells stimulates tumor cell growth through the formyl peptide receptor 1. Am. J. Pathol. 179, 1504–1512. https://doi.org/10.1016/j.ajpath.2011.05.059 (2011).
Yang, J., Zhang, L., Yu, C., Yang, X.-F. & Wang, H. Monocyte and macrophage differentiation: Circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark. Res. 2, 1. https://doi.org/10.1186/2050-7771-2-1 (2014).
Shi, Y. et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat. Commun. 8, 15080. https://doi.org/10.1038/ncomms15080 (2017).
Zhang, L. & Dimberg, A. Pleiotrophin is a driver of vascular abnormalization in glioblastoma. Mol. Cell Oncol. 3, e1141087. https://doi.org/10.1080/23723556.2016.1141087 (2016).
Wei, X. et al. Angiopoietin-like protein 2 facilitates non-small cell lung cancer progression by promoting the polarization of M2 tumor-associated macrophages. Am. J. Cancer Res. 7, 2220–2233 (2017).
Wang, Y.-Y. et al. Visfatin enhances breast cancer progression through CXCL1 induction in tumor-associated macrophages. Cancers 12, 3526. https://doi.org/10.3390/cancers12123526 (2020).
Cho, D. I. et al. Antiinflammatory activity of ANGPTL4 facilitates macrophage polarization to induce cardiac repair. JCI Insight 4, e125437. https://doi.org/10.1172/jci.insight.125437 (2020).
Deficiency of angiopoietin‐like 4 enhances CD8+ T cell bioactivity via metabolic reprogramming for impairing tumour progression—Ding—2023—Immunology. https://doi.org/10.1111/imm.13650. Accessed 30 Aug 2023.
Son, Y. M. et al. Resistin enhances the expansion of regulatory T cells through modulation of dendritic cells. BMC Immunol. 11, 33. https://doi.org/10.1186/1471-2172-11-33 (2010).
Sudan, S. K. et al. Resistin: An inflammatory cytokine with multi-faceted roles in cancer. Biochim. Biophys. Acta Rev. Cancer 1874, 188419. https://doi.org/10.1016/j.bbcan.2020.188419 (2020).
Hazini, A., Fisher, K. & Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother. Cancer 9, e002899. https://doi.org/10.1136/jitc-2021-002899 (2021).
Marchesi, M. et al. HLA-dependent tumour development: a role for tumour associate macrophages?. J. Transl. Med. 11, 247. https://doi.org/10.1186/1479-5876-11-247 (2013).
The NKG2A–HLA-E Axis as a Novel Checkpoint in the Tumor Microenvironment. Clinical Cancer Research. American Association for Cancer Research. https://aacrjournals.org/clincancerres/article/26/21/5549/82902/The-NKG2A-HLA-E-Axis-as-a-Novel-Checkpoint-in-the. Accessed 30 Aug 2023.
Brightman, S. E. et al. Tumor cells fail to present MHC-II–restricted epitopes derived from oncogenes to CD4+ T cells. JCI Insight 8, e165570. https://doi.org/10.1172/jci.insight.165570 (2020).
Mathewson, N. D. et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single cell analysis. Cell 184, 1281-1298.e26. https://doi.org/10.1016/j.cell.2021.01.022 (2021).
Shang, E. et al. Overexpression of CD99 is associated with tumor adaptiveness and indicates the tumor recurrence and therapeutic responses in gliomas. Transl. Oncol. 37, 101759. https://doi.org/10.1016/j.tranon.2023.101759 (2023).
Halliday, N. et al. CD86 Is a selective CD28 ligand supporting FoxP3+ regulatory T cell homeostasis in the presence of high levels of CTLA-4. Front. Immunol. 11, 600000. https://doi.org/10.3389/fimmu.2020.600000 (2020).
Frederico, S. C. et al. Making a cold tumor hot: The role of vaccines in the treatment of glioblastoma. Front. Oncol. 11, 672508. https://doi.org/10.3389/fonc.2021.672508 (2021).
Land, C. A., Musich, P. R., Haydar, D., Krenciute, G. & Xie, Q. Chimeric antigen receptor T-cell therapy in glioblastoma: charging the T cells to fight. J. Transl. Med. 18, 428. https://doi.org/10.1186/s12967-020-02598-0 (2020).
Author information
Authors and Affiliations
Contributions
A.J.T. and K.A.H. conceived the project and supervised the analyses. S.B. performed and supervised the computational analyses. S.B., K.A.H., N.R., S.S.G., and A.J.T. interpreted results and contributed to writing and editing the manuscript. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Batchu, S., Hanafy, K.A., Redjal, N. et al. Single-cell analysis reveals diversity of tumor-associated macrophages and their interactions with T lymphocytes in glioblastoma. Sci Rep 13, 20874 (2023). https://doi.org/10.1038/s41598-023-48116-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-48116-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.