Abstract
The Neotropical monophyletic catfish genus Harttia represents an excellent model to study karyotype and sex chromosome evolution in teleosts. Its species split into three phylogenetic clades distributed along the Brazilian territory and they differ widely in karyotype traits, including the presence of standard or multiple sex chromosome systems in some members. Here, we investigate the chromosomal rearrangements and associated synteny blocks involved in the origin of a multiple X1X2Y sex chromosome system present in three out of six sampled Amazonian-clade species. Using 5S and 18S ribosomal DNA fluorescence in situ hybridization and whole chromosome painting with probes corresponding to X1 and X2 chromosomes of X1X2Y system from H. punctata, we confirm previous assumptions that X1X2Y sex chromosome systems of H. punctata, H. duriventris and H. villasboas represent the same linkage groups which also form the putative XY sex chromosomes of H. rondoni. The shared homeology between X1X2Y sex chromosomes suggests they might have originated once in the common ancestor of these closely related species. A joint arrangement of mapped H. punctata X1 and X2 sex chromosomes in early diverging species of different Harttia clades suggests that the X1X2Y sex chromosome system may have formed through an X chromosome fission rather than previously proposed Y-autosome fusion.
Similar content being viewed by others
Introduction
Teleost fishes display an astounding variety of sex determination systems, involving genetic and environmental mechanisms or a combination thereof1,2,3. These mechanisms may largely differ among fish lineages and even among populations of the same species4,5,6,7. Particularly genetic sex determination is in fishes mainly governed by one of the nine presently known sex chromosome systems which evolved independently multiple times in different lineages6,7,8,9 and carry different sex-determining genes2,10. The majority of these systems show little genetic differentiation8,11,12 and are therefore prone to frequent sex chromosome turnovers7,13,14,15. These properties make teleost sex chromosomes a well-suited model for studying early phases of sex chromosome differentiation16,17 and causes and consequences of sex chromosome turnovers (whereby the newly evolved system replaces the former one)15,18,19, and the bearing of sex chromosome evolution to the establishment of reproductive barriers between incipient species20,21,22.
The study of sex chromosomes has undergone a remarkable transformation in recent years as genome sequencing, assembly and scaffolding techniques rapidly improved. Despite these advances, several unique biological features of sex chromosomes are still hardly tractable by computational tools, or their analysis requires multiple integrated methodologies and/or large number of individuals to be analyzed18,23. Cytogenetics and particularly the use of whole chromosome painting (WCP) probes enables comparative study among multiple (closely) related species, and it can be narrowed down specifically to linkage group(s) representing sex chromosomes8,24. This approach enables to determine whether sex chromosomes originated independently from different linkage groups or are formed by the same synteny blocks. The latter situation points either on a single shared origin of sex chromosomes or repeated and independent co-option of the same synteny blocks for the sex-determining role8,9,10. The use of cytogenetics may also avoid misinterpretations related to accidental involvement of sex-reversed individuals or the intra-specific variability in the sex-determining systems8,24.
The Neotropical armored catfish genus Harttia (Siluriformes, Loricariidae, Loricariinae) presently harbors 28 valid species25,26,27 together with three Harttia spp. determined based on cytogenetic features but waiting for a proper taxonomic description28. After Rineloricaria (reviewed in29), Harttia displays the second-largest variation in diploid chromosome number (2n) among Loricariidae fishes, ranging from 2n = 52♀/53♂ in H. carvalhoi30, to 2n = 62♀♂ in H. absaberi31 and Harttia sp. 232. Furthermore, the following three male-heterogametic sex chromosome systems have been identified in a subset of surveyed species: XX/XY1Y2 in H. carvalhoi, H. intermontana, and Harttia sp. 130,32,33; X1X1X2X2/X1X2Y in H. duriventris, H. punctata, and H. villasboas34,35, and a putative XX/XY in H. rondoni35. Together with African Nothobranchius killifishes36, Harttia represents a genus with the highest incidence of multiple sex chromosomes among teleosts to date8,32. It is therefore highly informative lineage for investigating underlying evolutionary forces that drive transitions from standard sex chromosomes (or other forms of sex determination) to multiple sex chromosome systems.
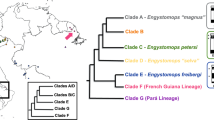
The most updated phylogeny of Loricariinae37, though not including all valid species, recognized the monophyly of Harttia, with the occurrence of three distinct evolutionary lineages: Clade I is composed of the species from Guyanese shield, Clade II includes the species from the Amazonian and Tocantins-Araguaia river basins, and Clade III harbors the species from the southern/southeastern Brazil. When complemented with a species set from a former phylogenetic study38 it is clear that Harttia species with known sex chromosomes, though nested within species lacking them based solely on a cytogenetic evidence, are grouped according to the type of their sex chromosome systems: those with an X1X1X2X2/X1X2Y system or tentative XX/XY sex chromosomes are placed in the clade II, and those with an XX/XY1Y2 system in the clade III28,32.
In our former studies, we demonstrated by WCP probes used in cross-species experiments (Zoo-FISH) that X1X2Y and XY1Y2 sex chromosome systems represent different linkage groups and therefore evolved independently39,40,41. While we also revealed by the same method the full or partial homeology between XY1Y2 systems among the three Harttia spp.39,41, similar information is lacking for XY and X1X2Y systems as yet. Indirect evidence based on ribosomal DNA (rDNA) physical mapping and comparative genomic hybridization (CGH) suggested that these systems might be potentially homeologous35. In this study, we aim to investigate the mechanism(s) of origin and the relationships between the sex chromosome systems in Harttia species belonging to the Amazonian clades I and II where three members are known to carry an X1X2Y sex chromosome system and a single species possesses putative XY sex chromosomes. We therefore probed altogether six related Harttia species with WCP probes derived from the X1 and X2 sex chromosomes of the X1X2Y system in H. punctata thus complementing our former study40. The analysis in the present study was complemented by mapping of rDNA clusters as these usually locate on Harttia sex chromosomes34,35,42. Our data show homeology between X1X2Y sex chromosome systems and also the putative XY sex chromosome system of H. rondoni. Among the two formerly proposed hypotheses on X1X2Y sex chromosome origin i.e. the Y-autosome fusion34,40 and X fission35, our results support the latter scenario.
Results
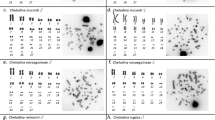
Cross-hybridization with HPU-X1 and HPU-X2 painting probes revealed full homeology between X1X2Y sex chromosome systems of H. punctata (analyzed by us formerly40), H. duriventris and H. villasboas (Fig. 1a,c). In H. duriventris and H. villasboas the HPU-X1 probe entirely hybridized to X2 chromosome and conversely, HPU-X2 probe painted X1 chromosome when following the nomenclature by Sassi et al.35. As the location of sex-determining region has not been identified yet, the assignation of X1 (ancestral) and X2 (neo) sex chromosomes in different species was done arbitrarily in former studies34,35. Hence, to avoid confusion in designation of demonstrably the same synteny blocks, we unified the nomenclature of sex chromosomes according to X1X2Y system of H. punctata34,40.
Zoo-FISH with HPU-X1 and HPU-X2 painting probes in male (first and third column) and female (second and fourth column) mitotic metaphases of Harttia duriventris (a), H. dissidens (d), H. rondoni (b), H. guianensis (e) H. villasboas (c), and Harttia sp. 3 (f). Chromosomes bearing 5S (red) and 18S (green) rDNA clusters as revealed after reprobing in the second FISH round are highlighted in boxes. The assignment of signals to specific chromosome pairs was performed based on data in our previous studies28,35. Full metaphase images are provided in Supplementary Fig. 1. Chromosomes were counterstained with DAPI (blue). Bar 10 µm.
The painting probes also labelled different portions of putative XY sex chromosome in H. rondoni (Fig. 1b) whose identity was confirmed by 18S rDNA mapping (see below). While the HPU-X1 probe painted the long (q) arms of these supposed X and Y chromosomes, the HPU-X2 probe painted short (p) arms of them both. Remarkably, a (peri)centric region of both chromosomes was left unstained by these probes. For the species without cytologically distinguishable sex chromosomes—H. dissidens H. guianensis, and Harttia sp. 3 (Fig. 1d–f) both painting probes, again, hybridized to a single metacentric chromosome pair. In H. dissidens and H. guianensis the hybridization pattern was the same as in H. rondoni (i.e. HPU-X1 covered q-arms while HPU-X2 stained p-arms of the fifth and fourth chromosome pair in H. dissidens and H. guianensis, respectively; compare Fig. 1d,e with Fig. 1b) but this time without the unstained region in (peri)centromeres. In Harttia sp. 3 (Fig. 1f) the painted chromosome corresponded to the 18S rDNA-bearing chromosome pair 1, with the HPU-X1 probe hybridizing on its p-arms and the HPU-X2 probe on its q-arms (i.e. the opposite scenario to the one found in H. rondoni, H. dissidens and H. guianensis). The (peri)centromeric region of this chromosome pair was left unstained by the painting probes.
18S rDNA signals were placed in the pericentromeric regions of X2 and Y chromosomes of H. villasboas and H. duriventris (Supplementary Fig. 1). These clusters also occupied (peri)centromeric positions in putative XY sex chromosomes in H. rondoni. Corroborating the previous study35, 18S rDNA cluster showed size heteromorphism with the X-linked site being notably extended compared to the Y-linked one. 18S rDNA probe further co-localized with the painting probes only in Harttia sp. 3 where it revealed a site on both homologs positioned on q-arms closely downstream of the centromere. Finally, in H. dissidens and H. guianensis, we observed a single pair of 18S rDNA-bearing chromosomes (pair 24 and 25, respectively, according to Sassi et al.28), with the centromere-proximal signal on the q-arms. In H. guianensis, the 18S rDNA-bearing chromosome pair also bore 5S rDNA cluster at the terminal portion of q-arms. Second pair of 5S rDNA signals in this species was located interstitially on the p-arms of metacentric chromosome pair 4. The same site was present also on chromosome pair 4 with the same morphology in H. rondoni where no additional 5S rDNA signals were detected. H. villasboas, H. duriventris and Harttia sp. 3 exhibited a single chromosome pair (no. 15) bearing 5S rDNA arrays on its p-arms. The same pattern but on chromosome pair 19 was found in H. dissidens.
All hybridization patterns are summarized in an ideogram (Fig. 2). Full metaphase images with rDNA hybridization patterns are provided in the Supplementary Fig. 1.
Schematic representation of hybridization results on chromosomes of studied Harttia species. For Harttia punctata the hybridization pattern was adopted from our previous study utilizing the same probes40. Letters correspond to those on Fig. 1. The assignment of signals to specific chromosome pairs, as well as the arrangement of chromosomes into a karyotype, was performed based on data in our previous studies28,35.
Discussion
We have shown herein by Zoo-FISH that Harttia species with X1X2Y sex chromosome system entirely share the synteny blocks by which these sex chromosomes are formed. Our findings also corroborate the existence of previously proposed35 XY sex chromosome system in H. rondoni, as also these chromosomes were stained by the same sex chromosome-derived painting probes.
Since the identification of the multiple XX/XY1Y2 sex chromosome system in H. carvalhoi30, Harttia catfish genus became an excellent model for studying sex chromosome evolution in Neotropical fishes, with three species having the X1X2Y sex chromosome system, other three the XY1Y2 one, and yet one another representative featuring tentative XY sex chromosomes. A series of cytogenetic studies relying mostly on repetitive DNA mapping, CGH and Zoo-FISH experiments provided already evidence that XY1Y2 systems found in H. carvalhoi, H. intermontana and Harttia sp. 1 are fully or partially homeologous among each other but are non-homologous to the X1X2Y system of H. punctata28,32,34,35,39,40,41,42. Homeology between X1X2Y sex chromosomes in H. punctata, H. duriventris and H. villasboas, and their close relationship to putative XY sex chromosomes in H. rondoni were previously proposed based on the shared presence of 18S rDNA clusters and CGH patterns35.
Blanco et al.34 initially hypothesized that the X1X2Y sex chromosomes in H. punctata originated from a Robertsonian translocation between the two acrocentic chromosomes—an ancestral Y and an autosome. This rearrangement would be accompanied by the loss of 18S rDNA arrays from the emerging neo-Y chromosome. Nonetheless, the chromosome painting data from the work by Deon et al.39 and our present study, once anchored to the current phylogenetic analysis (37Fig. 3A, 38Fig. 3B), clearly show that the linkage groups representing X1 and X2 chromosomes were ancestrally forming arms of the same chromosome. Besides the early diverging H. guianensis, this pattern has been found also in other Harttia lineages (39Fig. 3). This means that more probably the X1X2Y system emerged after a centric fission in the ancestral X chromosome.
Phylogenetic relationships among Hartiini fishes based on morphological and molecular data along with anchored cytogenetic characteristics. Phylogeny follows37 (a), and38 (b), while (c) presents the chromosomal data from species that were not included in the respective phylogenetic reconstructions. Indicated cytogenetic characteristics: 2n; sex chromosome systems; partial ideograms represent the organization of mapped synteny blocks as revealed by the sex chromosome-derived painting probes HPU-X1 (red) and HPU-X2 (green); 5S rDNA (blue), and 18S rDNA (black) sites. The assignment of signals to specific chromosome pairs was performed based on data in our previous studies28,32,35,40. More specifically, the cytogenetic data synthesis has been undertaken as follows: H. guianensis, H. dissidens and Harttia sp. 3 (28this study); H. punctata, H. kronei, H. loricariformis, H. longipinna, H. carvalhoi, H. torrenticola and H. gracilis40; H. duriventris, H. villasboas and H. rondoni (35this study); H. intermontana, Harttia sp. 1 and Harttia sp. 232,40; H. absaberi31.
When a fission event creates an X1X2Y multiple sex chromosome system, a closely related species carrying the ancestral XY sex chromosomes is expected to have a lower 2n in the karyotype43. This is what can be inferred from the comparison of H. rondoni with 2n = 54 (XY/XX) and the species with multiple X1X2Y/♀X1X1X2X2 sex chromosomes: H. punctata with (2n = 57♂/58♀), H. villasboas and H. duriventris (2n = 55♂/56♀).
CGH analysis has formerly shown that a probable region of differentiation on the Y chromosome might be located proximally to the centromere28. Indeed, the centromeric regions have been widely shown to suppress recombination44 and therefore are thought to be suitable regions for establishment of a new sex-determining region17. It is further intriguing that H. villasboas and H. duriventris share the presence of centromere-proximal 18S rDNA site on their Y chromosomes and that both XY chromosomes in H. rondoni share a pericentromeric 18S rDNA cluster being consistently larger on the X chromosome (35this study). It is tempting to hypothesize that the ancestral situation would be close to the scenario found in H. rondoni and the X-linked amplified 18S rDNA region might cause instability around the centromere, leading eventually to the fission, while selection would counteract similar fission to happen on the Y chromosome, to preserve the linkage disequilibrium in/around the sex-determining region.
rDNA clusters have been abundantly shown to cause chromosomal instability due to heavy transcription and organization into long tandem arrays45. More specifically, the following conditions collectively provide ample opportunities for DNA damage to happen: (1) highly decondensed DNA, (2) exposure of non-templated DNA strand and its tendency to form various secondary structures and (3) increased probability of collision between transcription and replication machineries. Consequent DNA repair may accidentally lead to rearrangements46,47,48. In the frame of the X-fission scenario, loss of rDNA sequences is among the possible consequences of double-stranded breaks in these tandem repeats49. Moreover, fission itself may lead to a partial degradation of exposed chromosomal ends until new telomeres are being established50. rDNA dynamics in Harttia is further evidenced by a complete loss of rDNA sites on certain linkage groups and their emergence on another chromosome pairs (40this study—see Fig. 2). Notably, besides Harttia spp.32,40, rDNA sites operated as breakpoint regions independently in many fish groups51,52,53,54,55.
While H. rondoni has not been involved in the current phylogenetic analysis37, former work38 proposed the phylogenetic position of this species being nested within the species carrying X1X2Y sex chromosomes, which might either mean that (1) ancestral XY system has been preserved in H. rondoni while X1X2Y system evolved repeatedly in separate evolutionary events in the closely related species or (2) there was a single origin of X1X2Y system and X1 and X2 fused secondarily back again in H. rondoni creating a neo-XY system, or (3) the phylogenetic relationships of H. rondoni with closely related species are not interpreted correctly by Covain et al.38. Regarding the last point, although geographical distribution and morphological characters reinforce the Covain’s proposition25, further phylogenetic studies involving the species in question are necessary to untangle this issue. Noteworthily, if X1X2Y sex chromosomes evolved multiple times independently in this Harttia lineage, then the repeated fusion of ancestral Y with always the same autosome is rather improbable9,56,57,58, which, again, reinforces the X-fission hypothesis.
The origin of multiple X1X2Y sex chromosome system in the three Harttia species seems not to follow the common evolutionary pathway. A centric or tandem fusion of the original Y chromosome with an autosome has been proposed (and in several cases empirically confirmed) as an underlying mechanism leading to emergence of X1X2Y sex chromosomes in the remaining 62 teleosts cases reported to date8,55,59. It is also the commonest mechanism of multiple sex chromosome creation in other cold-blooded vertebrates60,61. Sex chromosome fissions are much less common and in teleosts they have been proposed thus far only in five cases (four times as Y-fission and once as W-fission; reviewed in8). Another tentative W-fission might have taken place in Ancistrus clementinae62. Finally, a Y-fission has been proposed also for the XY1Y2 system in H. carvalhoi33, however, a more recent study32 suggested X-autosome fusion to be responsible instead, which has been further reinforced by Zoo-FISH showing that a probable ancestral X chromosome fused with two different autosomes within the set of three XY1Y2-bearing Harttia species39.
Fissions are generally hard to track in the species' karyotypes as they generate less noticeable products compared to large chromosomes derived from fusions50,63. Hence, unfortunately, our understanding of the genomic properties of fission sites and the etiology of this rearrangement type are still rather limited, despite the steadily growing number of sequenced genomes in non-model organisms. Studies combining chromosomal painting with specific sex-chromosome probes and other cytogenetic markers, such as the distribution of repetitive DNAs, are useful to indicate major fission events, as already proven in fishes64,65, lizards56,66, and birds67. Such an approach is desirable as fissions have been associated with chromosomal evolution and speciation events in several metazoans68,69,70 and they have been also, for instance, associated with the evolution of the olfactory system in carnivores71.
While sex chromosome-autosome fusions have been much more explored regarding their possible effects on reproductive isolation, adaptation and radiation8,20,57,60 fissions might have a similar effect72. In the case of Harttia species, given that these fishes form rather small, fragmented populations with restricted/absent gene flow, multiple sex chromosomes might have emerged and get fixed rather under the major effect of genetic drift73 which highly likely applies also to several other Neotropical fishes with multiple sex chromosomes55,65,74. The possible contribution of natural selection in this process will require further studies particularly oriented towards a detailed characterization of genetic content of sex-determining regions and neighboring chromosomal areas.
Conclusion
The chromosomal evolution in Harttia species has been shaped by numerous inter-chromosomal rearrangements giving rise to a complex karyotype variability, including the emergence of different male-heterogametic sex chromosome systems. In this study, we demonstrated that the X1X2Y and XY sex chromosomes, as well as some autosomes, share several homologies among Harttia species. This strengthens our previously proposed theory that the X1X2Y system emerged after a centric fission in the ancestral X chromosome and thus represents a derivation of the ancestral XY sex chromosome system. Although the amount of chromosomal data has significantly increased for Harttia species during the recent years, the genus still lacks a robust phylogenetic reconstruction that would include all recognized species along with the other emerging but yet undescribed species whose existence has been proposed by their distinct cytogenetic features28.
Methods
Sampling and chromosome preparation
Species were gathered from seven distinct localities (Fig. 4, Table 1), with the authorization of the Brazilian environmental agency ICMBIO/SISBIO (License 48628-14) and SISGEN (A96FF09). Mitotic chromosomes were obtained by the classic air-drying method75, using the anterior kidney cells as the main source material, being occasionally supplemented with the cells of the spleen tissue. All procedures followed the ethical and anesthesia conducts approved by the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315). The authors complied with ARRIVE guidelines. Specimens were fixed in 10% formalin and deposited in the fish collections of the Instituto Nacional de Pesquisa da Amazônia (INPA-ICT) and Museu de Zoologia da Universidade de São Paulo (MZUSP). Their voucher numbers are provided in Table 1.
Partial map of Brazil highlighting the Amazonian (green) and Tocantins-Araguaia (orange) river basins. Circles correspond to sampling sites of Harttia species whose sex chromosome systems are indicated by specific colors. The color-coding system for river basins and sex chromosomes is presented in frames (bottom right). 1 = H. guianensis; 2 = H. dissidens; 3 = H. duriventris; 4 = H. rondoni; 5 = H. villasboas; 6 = H. punctata and 7 = Harttia sp. 3. The map was created with QGIS 3.22 with the package Natural Earth.
Probe preparation for Zoo-FISH and rDNA FISH
Sex chromosomes of H. punctata were selected to be used as probes since this species exhibits the closest 2n relative to the proposed ancestral state for the genus and, at the same time, it possesses a multiple sex chromosome system of the X1X1X2X2/X1X2Y type28,32,34,35,42. Fifteen copies of the X1 and X2 chromosomes were isolated by glass-needle-based microdissection under an inverted microscope (Zeiss Axiovert 135). The collected DNA material was then amplified in a primary degenerated oligonucleotide-primed polymerase chain reaction (DOP-PCR)76. The probes were then labeled in the secondary DOP-PCR reaction using 1 µL of the initial amplified product as a template DNA77. The probe derived from the X1 chromosome (HPU-X1) was labeled with Spectrum Orange-dUTP (red), and the one derived from the X2 chromosome (HPU-X2) with Spectrum Green-dUTP (green) (Vysis, Downers Grove, United States).
The 5S and 18S rDNA fragments were obtained by PCR from the wolf fish Hoplias malabaricus genome using primers and thermal profiles described in previous studies78,79,80. The labelling was done by nick translation using Atto550-dUTP (red) for the 5S rDNA and Alexa Fluor 488-dUTP (green) for the 18S rDNA (both Jena Biosciences, Jena, Germany), according to manufacturer's protocol.
FISH experiments
Zoo-FISH followed the protocol described by Sassi et al.81. C0t-1 DNA prepared from H. punctata male genome was used as a blocker to high-copy repeat sequences82. Slides with metaphase chromosomes of H. dissidens, H. duriventris, H. guianensis, H. villasboas, H. rondoni and Harttia sp. 3 were denatured in 70% formamide/2 × SSC at 72 °C for 3 min. For each assay, the hybridization solution (200 ng of HPU-X1, 200 ng of HPU-X2 and 20 µg of C0t-1 DNA in 50% formamide + 2 × SSC + 10% dextran sulfate; final volume 20 µL) was denatured for 10 min at 85 °C, cooled at 4 °C for 2 min, and allowed to pre-anneal for 45 min at 37 °C in a thermocycler. Next, the probes were spotted onto the denatured slides, and the hybridization process took place in a dark moist chamber for 48 h at 37 °C. To remove unspecific hybridization signals, slides were washed twice with 1 × SSC at 65 °C (5 min each), and then in 4 × SSC/Tween (5 min) and 1 × PBS (1 min), at room temperature. After capturing the resulting images, the slides were washed for the second round of hybridization80, in which 100 ng of each 5S and 18S rDNA probe was applied after being dissolved in the hybridization solution (50% formamide and 10% dextran sulfate in 2 × SSC), in the final volume 20 µL. The rDNA FISH experiments followed the same protocol as described above, except for the hybridization time which was 24 h. In all experiments, chromosomes were finally counterstained with VECTASHIELD® Antifade Mounting Medium with DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories, California, United States).
Microscopy and image analysis
Hybridization patterns were verified in at least 30 metaphase spreads per experiment. Images were captured using an Olympus BX50 microscope (Olympus Corporation, Ishikawa, Japan), coupled with a CoolSNAP camera. Images were processed with Ikaros/ISIS (MetaSystems, Germany). The assignment of signals to specific chromosome pairs, as well as the arrangement of chromosomes into a karyotype, was performed based on data in our previous studies28,35.
Ethical approval
Sample was approved by the Brazilian Environmental Agency ICMBIO/SISBIO (License 48628-14) and SISGEN (A96FF09). All experiments followed the guidelines and were approved by the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315 and 7994170423).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Devlin, R. H. & Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 208, 191–364 (2002).
Guiguen, Y., Fostier, A. & Herpin, A. Sex determination and differentiation in fish: Genetic, genomic, and endocrine aspects. In Sex Control in Aquaculture (eds Wang, H. P. et al.) 35–63 (Wiley, 2019). https://doi.org/10.1002/9781119127291.ch2.
Shen, Z. G. & Wang, H. P. Environmental sex determination and sex differentiation in teleosts—how sex is established. In Sex Control in Aquaculture (eds Wang, H. P. et al.) 85–115 (Wiley, 2019). https://doi.org/10.1002/9781119127291.ch4.
Wilson, C. A. et al. Wild sex in zebrafish: Loss of the natural sex determinant in domesticated strains. Genetics 198, 1291–1308 (2014).
Yamamoto, Y., Zhang, Y., Sarida, M., Hattori, R. S. & Strüssmann, C. A. Coexistence of genotypic and temperature-dependent sex determination in pejerrey Odontesthes bonariensis. PLoS One 9, e102574 (2014).
Myosho, T., Takehana, Y., Hamaguchi, S. & Sakaizumi, M. Turnover of sex chromosomes in celebensis group medaka fishes. G3 (Bethesda) 5, 2685–2691 (2015).
El Taher, A., Ronco, F., Matschiner, M., Salzburger, W. & Böhne, A. Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci. Adv. 7, eabe8215 (2021).
Sember, A. et al. Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: State of the art and future challenges. Philos. Trans. R. Soc. B 376, 20200098 (2021).
Jeffries, D. L., Mee, J. A. & Peichel, C. L. Identification of a candidate sex determination gene in Culaea inconstans suggests convergent recruitment of an Amh duplicate in two lineages of stickleback. J. Evol. Biol. 35, 1683–1695 (2022).
Pan, Q. et al. Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Philos. Trans. R. Soc. B Biol. Sci. 376, 20200091 (2021).
Gamble, T. Using RAD-seq to recognize sex-specific markers and sex chromosome systems. Mol. Ecol. 25, 2114–2116 (2016).
Schartl, M., Schmid, M. & Nanda, I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma 125, 553–571 (2016).
Mank, J. E. & Avise, J. C. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex. Dev. 3, 60–67 (2009).
Kabir, A. et al. Repeated translocation of a supergene underlying rapid sex chromosome turnover in Takifugu pufferfish. Proc. Natl. Acad. Sci. USA A119, e2121469119 (2022).
Vicoso, B. Molecular and evolutionary dynamics of animal sex-chromosome turnover. Nat. Ecol. Evol. 3, 1632–1641 (2019).
Kamiya, T. et al. A Trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (Fugu). PLoS Genet. 8, e1002798 (2012).
Charlesworth, D. Young sex chromosomes in plants and animals. New Phytol. 224, 1095–1107 (2019).
Palmer, D. H., Rogers, T. F., Dean, R. & Wright, A. E. How to identify sex chromosomes and their turnover. Mol. Ecol. 28, 4709–4724 (2019).
Saunders, P. A. Sex chromosome turnovers in evolution. eLS 20, 1–8. https://doi.org/10.1002/9780470015902.a0028747 (2019).
Kitano, J. et al. A role for a neo-sex chromosome in stickleback speciation. Nature 461, 1079–1083 (2009).
O’Neill, M. J. & O’Neill, R. J. Sex chromosome repeats tip the balance towards speciation. Mol. Ecol. 27, 3783–3798 (2018).
Payseur, B. A., Presgraves, D. C. & Filatov, D. A. Introduction: Sex chromosomes and speciation. Mol. Ecol. 27, 3745–3748 (2018).
Carey, S. B. et al. Representing sex chromosomes in genome assemblies. Cell Genom. 2, 100132 (2022).
Deakin, J. E. et al. Chromosomics: Bridging the gap between genomes and chromosomes. Genes 10, 627 (2019).
Oyakawa, O. T., Fichberg, I. & Py-Daniel, L. R. Three new species of Harttia (Loricariidae: Loricariinae) from Serra do Cachimbo, Rio Xingu basin, Pará, Northern Brazil. Zootaxa 4387, 75–90 (2018).
Caldas, L., Cherobim, A. M. & Langeani, F. A New species of Harttia from the rio São Francisco basin (Siluriformes: Loricariidae). Neotrop. Ichthyol. 20, 25 (2022).
Fricke, R., Eschmeyer, W. N. & van der Laan, R. Eschmeyer’s catalog of fishes: Genera, Species, References. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (2023).
Sassi, F. M. C. et al. Adding new pieces to the puzzle of karyotype evolution in Harttia (Siluriformes, Loricariidae): Investigation of Amazonian species. Biology 10, 922 (2021).
Takagui, F. H. et al. Unrevealing the karyotypic evolution and cytotaxonomy of armored catfishes (Loricariinae) with emphasis in Sturisoma, Loricariichthys, Loricaria, Proloricaria, Pyxiloricaria, and Rineloricaria. Zebrafish 17, 319–332 (2020).
Centofante, L., Bertollo, L. A. C. & Moreira-Filho, O. Cytogenetic characterization and description of an XX/XY1Y2 sex chromosome system in catfish Harttia carvalhoi (Siluriformes, Loricariidae). Cytogenet. Genome Res. 112, 320–324 (2006).
Rodrigues, R. M. Estudos cromossômicos e moleculares em Loricariinae com ênfase em espécies de Rineloricaria (Siluriformes, Loricariidae): Uma perspectiva evolutiva (Ph.D. Thesis, Universidade de São Paulo, 2010).
Deon, G. A. et al. Highly rearranged karyotypes and multiple sex chromosome systems in armored catfishes from the genus Harttia (Teleostei, Siluriformes). Genes 11, 1366 (2020).
Blanco, D. R. et al. The role of the Robertsonian rearrangements in the origin of the XX/XY1Y2 sex chromosome system and in the chromosomal differentiation in Harttia species (Siluriformes, Loricariidae). Rev. Fish Biol. Fish. 23, 127–134 (2013).
Blanco, D. R. et al. Origin of the X1X1X2X2/X1X2Y sex chromosome system of Harttia punctata (Siluriformes, Loricariidae) inferred from chromosome painting and FISH with ribosomal DNA markers. Genetica 142, 119–126 (2014).
Sassi, F. M. C. et al. Multiple sex chromosomes and evolutionary relationships in amazonian catfishes: The outstanding model of the genus Harttia (Siluriformes: Loricariidae). Genes 11, 1179 (2020).
Krysanov, E. & Demidova, T. Extensive karyotype variability of African fish genus Nothobranchius (Cyprinodontiformes). Comp. Cytogenet. 12, 387 (2018).
Londoño-Burbano, A. & Reis, R. E. A combined molecular and morphological phylogeny of the Loricariinae (Siluriformes: Loricariidae), with emphasis on the Harttiini and Farlowellini. PLoS One 16, e0247747 (2021).
Covain, R. et al. Molecular phylogeny of the highly diversified catfish subfamily Loricariinae (Siluriformes, Loricariidae) reveals incongruences with morphological classification. Mol. Phylogenet. Evol. 94, 492–517 (2016).
Deon, G. A. et al. Chromosomal rearrangements and origin of the multiple XX/XY1Y2 sex chromosome system in Harttia species (Siluriformes: Loricariidae). Front. Genet. 13, 877522 (2022).
Deon, G. A. et al. Evolutionary breakpoint regions and chromosomal remodeling in Harttia (Siluriformes: Loricariidae) species diversification. Genet. Mol. Biol. 45, e20210170 (2022).
Sassi, F. M. C. et al. Turnover of multiple sex chromosomes in Harttia catfish (Siluriformes, Loricariidae): A glimpse from whole chromosome painting. Front. Genet. 14, 1226222 (2023).
Blanco, D. R. et al. Karyotype diversity and evolutionary trends in armored catfish species of the genus Harttia (Siluriformes: Loricariidae). Zebrafish 14, 169–176 (2017).
Kitano, J. & Peichel, C. L. Turnover of sex chromosomes and speciation in fishes. Environ. Biol. Fish. 94, 549–558 (2012).
Nambiar, M. & Smith, G. R. Repression of harmful meiotic recombination in centromeric regions. Mech. Cancer Cachexia 54, 188–197 (2016).
Schöfer, C. & Weipoltshammer, K. Nucleolus and chromatin. Histochem. Cell Biol. 150, 209–225 (2018).
Potapova, T. A. & Gerton, J. L. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res. 27, 109–127 (2019).
Warmerdam, D. O. & Wolthuis, R. M. F. Keeping ribosomal DNA intact: A repeating challenge. Chromosome Res. 27, 57–72 (2019).
Goffová, I. & Fajkus, J. The rDNA loci—intersections of replication, transcription, and repair pathways. Int. J. Mol. Sci. 22, 1302 (2021).
Warmerdam, D. O., van den Berg, J. & Medema, R. H. Breaks in the 45S rDNA lead to recombination-mediated loss of repeats. Cell Rep. 14, 2519–2527 (2016).
Perry, J., Slater, H. R. & Choo, K. H. A. Centric fission—simple and complex mechanisms. Chromosome Res. 12, 627–640 (2004).
Cioffi, M. B. & Bertollo, L. A. C. Initial steps in XY chromosome differentiation in Hoplias malabaricus and the origin of an X1X2Y sex chromosome system in this fish group. Heredity 105, 554–561 (2010).
Barros, A. V. et al. Fragile sites, dysfunctional telomere and chromosome fusions: What is 5S rDNA role?. Gene 608, 20–27 (2017).
Supiwong, W. et al. Karyotype diversity and evolutionary trends in the Asian swamp eel Monopterus albus (Synbranchiformes, Synbranchidae): A case of chromosomal speciation?. BMC Evol. Biol. 19, 73 (2019).
Sember, A. et al. Centric fusions behind the karyotype evolution of Neotropical Nannostomus pencilfishes (Characiforme, Lebiasinidae): First insights from a molecular cytogenetic perspective. Genes 11, 91 (2020).
Marajó, L. et al. Chromosomal rearrangements and the first indication of an ♀X1X1X2X2/♂X1X2Y sex chromosome system in Rineloricaria fishes (Teleostei: Siluriformes). J. Fish Biol. 102, 443–454 (2023).
Giovannotti, M. et al. New insights into sex chromosome evolution in anole lizards (Reptilia, Dactyloidae). Chromosoma 126, 245–260 (2017).
Carabajal Paladino, L. Z. et al. Sex chromosome turnover in moths of the diverse superfamily Gelechioidea. Genome Biol. Evol. 11, 1307–1319 (2019).
Oliveira da Silva, W. et al. Identification of two independent X-autosome translocations in closely related mammalian (Proechimys) species. Sci. Rep. 9, 4047 (2019).
Ferchaud, A.-L. et al. Chromosome-level assembly reveals a putative Y-autosomal fusion in the sex determination system of the Greenland Halibut (Reinhardtius hippoglossoides). G3 (Bethesda) 12, jkab376 (2022).
Pennell, M. W. et al. Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 11, e1005237 (2015).
Ma, W. J. & Veltsos, P. The diversity and evolution of sex chromosomes in frogs. Genes 12, 483 (2021).
Nirchio, M. et al. Occurrence of sex chromosomes in fish of the genus Ancistrus with a new description of multiple sex chromosomes in the Ecuadorian endemic Ancistrus clementinae (Loricariidae). Genes 14, 306 (2023).
Imai, H. T., Satta, Y. & Takahata, N. Integrative study on chromosome evolution of mammals, ants and wasps based on the minimum interaction theory. J. Theor. Biol. 210, 475–497 (2001).
Schemberger, M. O. et al. Differentiation of repetitive DNA sites and sex chromosome systems reveal closely related group in Parodontidae (Actinopterygii: Characiformes). Genetica 139, 1499–1508 (2011).
de Oliveira, E. A. et al. Tracking the evolutionary pathway of sex chromosomes among fishes: Characterizing the unique XX/XY1Y2 system in Hoplias malabaricus (Teleostei, Characiformes). Chromosoma 127, 115–128 (2018).
Gladkikh, O. L. et al. Rapid karyotype evolution in Lasiopodomys involved at least two autosome – sex chromosome translocations.PLoS ONE. 11, e0167653 (2016)
Kretschmer, R. et al. Extensive chromosomal fissions and repetitive DNA accumulation shaped the atypical karyotypes of two Ramphastidae (Aves: Piciformes) species. Biol. J. Linn. Soc. Lond. 130, 839–849 (2020).
Voss, S. R. et al. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. 21, 1306–1312 (2011).
de Vos, J. M., Augustijnen, H., Bätscher, L. & Lucek, K. Speciation through chromosomal fusion and fission in Lepidoptera. Philos. Trans. R. Soc. B Biol. Sci. 375, 20190539 (2020).
Huang, Z. et al. Recurrent chromosome reshuffling and the evolution of neo-sex chromosomes in parrots. Nat. Commun. 13, 944 (2022).
Fan, H. et al. Chromosome-level genome assembly for giant panda provides novel insights into Carnivora chromosome evolution. Genome Biol. 20, 267 (2019).
Yoshido, A. et al. Evolution of multiple sex-chromosomes associated with dynamic genome reshuffling in Leptidea wood-white butterflies. Heredity 125, 138–154 (2020).
Saunders, P. A., Neuenschwander, S. & Perrin, N. Sex chromosome turnovers and genetic drift: A simulation study. J. Evol. Biol. 31, 1413–1419 (2018).
de Souza, F. H. S. et al. Integrating cytogenetics and population genomics: Allopatry and neo-sex chromosomes may have shaped the genetic divergence in the Erythrinus erythrinus species complex (Teleostei, Characiformes). Biology 11, 315 (2022).
Bertollo, L. A. C., Cioffi, M. B. & Moreira-Filho, O. Direct chromosome preparation from freshwater teleost fishes. In Fish Cytogenetic Techniques (eds Ozouf-Costaz, C. et al.) 21–26 (CRC Press, 2015). https://doi.org/10.1201/b18534-4.
Yang, F., Trifonov, V., Ng, B. L., Kosyakova, N. & Carter, N. P. Generation of paint probes by flow-sorted and microdissected chromosomes. In Fluorescence In Situ Hybridization (FISH)—Application Guide (ed. Liehr, T.) 35–52 (Springer, 2009). https://doi.org/10.1007/978-3-540-70581-9_3.
Yang, F. & Graphodatsky, A. S. Animal probes and ZOO-FISH. In Fluorescence In Situ Hybridization (FISH)—Application Guide (ed. Liehr, T.) 323–346 (Springer, 2009). https://doi.org/10.1007/978-3-540-70581-9_29.
Pendás, A. M., Móran, P., Freije, J. P. & Garcia-Vásquez, E. Chromosomal location and nucleotide sequence of two tandem repeats of the Atlantic salmon 5S rDNA. Cytogenet. Cell Genet. 67, 31–36 (1994).
Cioffi, M. B., Martins, C., Centofante, L., Jacobina, U. & Bertollo, L. A. C. Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: Mapping of three classes of repetitive DNAs. Cytogenet. Genome Res. 125, 132–141 (2009).
Yano, C. F., Bertollo, L. A. C. & Cioffi, M. B. Fish-FISH: Molecular cytogenetics in fish species. In Fluorescence In Situ Hybridization (FISH)—Application Guide (ed. Liehr, T.) 429–444 (Springer, 2017).
Sassi, F. M. C., Toma, G. A. & Cioffi, M. B. FISH—in fish chromosomes. In Cytogenetics and Molecular Cytogenetics (ed. Liehr, T.) 281–293 (CRC Press, 2023).
Zwick, M. S. et al. A rapid procedure for the isolation of C0t–1 DNA from plants. Genome 40, 138–142 (1997).
Acknowledgements
The authors are grateful to Eliana Feldberg and Lúcia Helena Rapp Py-Daniel from National Institute of Amazonian Research who provided assist in the collection and identification of animal samples. We also thanks to FAPESP for the funding.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by São Paulo Research Foundation (FAPESP) grants 2020/02681-9, 2022/04261-2, and 2023/08116-0 (FMCS), 2020/11772-8 (MBC), Brazilian National Council for Scientific and Technological Development (CNPq), Grant number 302928/2021-9 (MBC) and Czech Academy of Sciences (RVO: 67985904 of IAPG CAS, Liběchov) (AS).
Author information
Authors and Affiliations
Contributions
F.M.C.S., G.A.D., O.M.F. and M.B.C. conceived and designed research. F.M.C.S. and G.A.D. conducted experiments. F.M.C.S., G.A.D., T.L., A.S., O.M.F., L.A.C.B., M.R.V., and M.B.C. analyzed the data. A.S., T.L., O.T.O. contributed with new methods. F.M.C.S., A.S., G.A.D., L.A.C.B., M.R.V., T.L. and M.B.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Menezes Cavalcante Sassi, F., Sember, A., Deon, G.A. et al. Homeology of sex chromosomes in Amazonian Harttia armored catfishes supports the X-fission hypothesis for the X1X2Y sex chromosome system origin. Sci Rep 13, 15756 (2023). https://doi.org/10.1038/s41598-023-42617-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42617-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.