Abstract
Chitinase-producing fungi have now engrossed attention as one of the potential agents for the control of insect pests. Entomopathogenic fungi are used in different regions of the world to control economically important insects. However, the role of fungal chitinases are not well studied in their infection mechanism to insects. In this study, Chitinase of entomopathogenic fungi Trichoderma longibrachiatum was evaluated to control Aphis gossypii. For this purpose, fungal chitinase (Chit1) gene from the genomic DNA of T. longibrachiatum were isolated, amplified and characterised. Genomic analysis of the amplified Chit1 showed that this gene has homology to family 18 of glycosyl hydrolyses. Further, Chit1 was expressed in the cotton plant for transient expression through the Geminivirus-mediated gene silencing vector derived from Cotton Leaf Crumple Virus (CLCrV). Transformed cotton plants showed greater chitinase activity than control, and they were resistant against nymphs and adults of A. gossypii. About 38.75% and 21.67% mortality of both nymphs and adults, respectively, were observed by using Chit1 of T. longibrachiatum. It is concluded that T. longibrachiatum showed promising results in controlling aphids by producing fungal chitinase in cotton plants and could be used as an effective method in the future.
Similar content being viewed by others
Introduction
Crops are vulnerable to attack by many biotic factors including phytopathogens1. Mostly, insect pests not only involved in yield losses but also affect the overall quality and quantity of agricultural products2. Aphids are the most devastating economical pest of various field, ornamental and greenhouse crops such as tomato, cucumber, pepper, cotton, wheat, brassica and rose3,4,5. Cotton (Gossypium hirsutum L.) is an important cash crop and is highly susceptible to attack by aphids6. Cotton aphids (Aphis gossypii), as a vicious pest of cotton that usually suck the phloem sap from vascular plant parts of cotton and help in the transmission mechanism of more than 75 viruses7,8. Major damages caused by A. gossypii are associated with its remarkable deposition of sooty mold and secretion of honeydews on cotton balls9,10. Cotton aphids areone of the notorious pests due to viviparous in nature and has high production rates in a short interval of oviposition periods9.
In the past, aphids were usually managed using various insecticides in both greenhouse and field crops; but it has resulted in severe socio-economical, environmental and health problems10. The high and injudicious uses of chemical insecticides have enhanced the resurgence and resistance of most insect pests. Besides this, the high input cost of insecticides and all legal restrictions have focused on using an alternative approach to control insect pests11,12. In the previous years, an alternative bio-control method is in specifically, the use of entomopathogens has emerged13,14. Entomopathogenic fungi [EPF] have exclusive characterisations that enable them to be the potential bio-control agents of insect pests. Due to their vast host range, almost 1000 different species of EPF are involved in effectively controlling many insects species15,16. Various insect species such as thrips, whiteflies, aphids, mealybugs, psyllids, weevils and nematodes are hosts of EPF in both field and greenhouses17,18,19. Different species and strains of Trichoderma like T. afroharzianum and T. atroviride are actively found in the bio-control of aphids at their different morphological stages. Studies carried out to date have reported that Trichoderma is capable of controlling insect pests directly through parasitism and by the production of insecticidal secondary metabolites, antifeedant compounds and repellent metabolites. While, indirectly through the activation of systemic plant defensive responses, the attraction of natural enemies or the parasitism of insect-symbiotic microorganisms. It has been shown that Trichoderma may also have complementary properties that enhance plant defense barriers against insect pests20,21. However, it is not easy to apply entomopathogens under field conditions along with other management practices due to differences in their mode of action22. Currently, scientists are applying biotechnological techniques to find unique genes that can be incorporated into the genome of the host plant to create resistance against specific insect pests. Over the past few years, various studies have highlighted the secretion of specific enzymes such as chitinases and proteases by EPF, which are playing a crucial role as insecticidal23. The outer cuticle of the insects is usually composed of chitin, embedded with a protein matrix. The extracellular enzymatic compounds produced by EPF can degrade these chitinous compounds and obtain nutrients from the insect’s hemolymph24,25. The fungal chitinases usually belong to class 3 chitinases and are considered an important part of the family 18 of glycosyl hydrolases26. Fungal chitinases of family 18 contain a chitin-binding domain that efficiently depolarises chitin27,28. The main structural component of an insect’s body is chitin that is a poly-β-1, 4-N-acetylglucosamine and almost 40% of the insect’s body mass is composed of it29,30. Taking into consideration, chitinase from the insect's associated fungi can be used to develop transgenic plants for the effective control of aphids. The overall expression of the Chit1 gene in T. longibrachiatum is actively involved in enhancing the efficiency of insect cuticle digestion and enabling the increased virulence against insects. The role of various fungal secondary metabolites and extracellular enzymes is well documented to control the population of aphids31,32.
Genetic engineering techniques are employed to develop resistance in cotton plants against fungal pathogens, but there is lack of data related to the transgenic approach of chitinase genes against insect pests33. Various research studies are in progress about the transgenic expression of chitinases of fungus in cotton for transgenic plant development34. Therefore, the main objectives of our study were to isolate and characterise the chitinase gene from the fungal strain of T. longibrachiatum, development of transgenic cotton plants and analyse the overall resistance of transgenics against cotton aphids. The projected results will help to understand the use of transient chitinase plants for the bio-control of insects pests in the future.
Results
Isolation of endochitinase Chit1
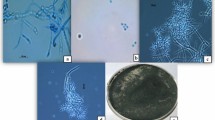
The chitinase gene was isolated from the genomic DNA of T. longibrachiatum and a product size of approximately 1 kb for Chit1 is produced through PCR (Fig. 1a). The restriction digestion resulted in confirmation of partial recombinant clones of Chit1 (Fig. 1b), and M13 primers were further used to sequence. The sequences of vector were removed from the nucleotide sequence using GENE TOOL and then subjected to BLAST analysis. The result confirms that the nucleotide sequences of Chit1 had a maximum resemblance to endochitinases as compared to other EPF. Chit1 showed remarkable homology of 99.52% to endochitinase from T. longibrachiatum Chit1 (KJ787131).
Characterisation of endochitinase Chit1 from an isolate of T. longibrachiatum
The endochitinase gene of T. longibrachiatum was mainly composed of 216 amino acids.. The molecular mass of the encoded protein of T. longibrachiatum was 23.41 kDa, acidic in nature with a pI value of 4.5 (Table 1). In the case of T. longibrachiatum, the gene's sequence analyses expressed its origin. It was assessed that this gene has a domain belonging to catalytic glycosyl hydrolase family 18. The sequence alignment explained about the structural motif. The catalytic domain DGIDVDWEYP (DxxDxDxE) is dependent on the structural motif and is conserved in the chitinase gene. The substrate's binding site was at 6aa, while the active site of the catalyst was placed at site 43aa. The sequence was blasted, and it showed 99% similarity with the sequence of T. longibrachiatum strain ACD46738 and CDM98718.
Phylogenetic analysis of T. longibrachiatum
The phylogenetic analysis was performed to find out the genetic variation among chitinases of different Trichoderma species. In phylogenetic tree analysis, endochitinase of fungal isolates were grouped into various clades and it showed maximum homologies to class 18 basic chitinases. T. longibrachiatum (Accession no: OQ689733) was classified into distant clades, and it showed maximum resemblance with T. longibrachiatum Chit1 (KJ787131). Overall comparative sequence analysis exposed the fact that endochitinase Chit1 is most related to the 18 glycosyl hydrolases family (Fig. 2).
Chitinase confirmation in cotton plants
To confirm the chitinases in cotton plants, ORF the encoding conserved domain of catalytic family 18 of Chit1 genes T. longibrachiatum was selected. The selection was made to detect the presence of the chitinases gene in cotton plants through a VIGS vector that is using a component of CLCrV. The amplicon of around 900 bp was observed from the amplification of recombinant plasmids using specific ORFs primers (Fig. 3a). VGS-Chit1_ Tl construct was confirmed by restriction analysis and sequencing (Fig. 3b). After 7 days of inoculation, amplification was positive for all plants. The development of different symptoms of CLCrV on cotton plants also revealed the expression of chitinase in plants as CLCrV is modified natural vector from Cotton Curl Crumple Virus and it shows similar symptoms in the expression of a particular gene (Fig. 4).
Chitinase activity assays
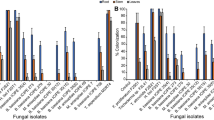
The transformed cotton plants produced chitinase after 5 days. All the plants, including mock and control had produced chitinase, but chitinase production is higher in transformed plants than that of control plants. The transformed cotton plants of Chit1 of T. longibrachiatum showed the chitinase activity of 5.64 U/mL. The overall difference in chitinase activity was found by reducing the chitinase gene from the control value. The chitinase activity of the control plant that had both VIGS and gene GUS was 5.09 U/mL, whereas chitinase activity was 5.44 U/mL in mock plants. The overall chitinase activity was shown in Table 2.
Bioassays of transformed cotton plants against nymphs and adults of A. gossypii
Transformed cotton plants exhibited excellent virulence against the 4th nymphal instar and adults of A. gossypii. Chit1 of T. longibrachiatum expressed in cotton plants showed promising results and the mortality of nymphs of about 38.75% after 144 h (Table 3). The highest adult mortality of 21.67% was recorded for Chit1 T. longibrachiatum transgenic plants (Table 3). The virulence efficiency of T. longibrachiatum can be increased by the overall production of chitinases, and the nymphal stage was more vulnerable to mortality compared to adults.
Discussion
Entomopathogenic fungi are considered a potential microbial bio-control agent due to the production of various secondary metabolites and enzymes that act as insecticide and bio-pesticides. Plants are vulnerable to attack by pathogens, and usually, various resistance genes are encoded in plants to impart resistance. Chitinase genes canbecome effective now–a–day just like Bacillus thuringiensis, Bt gene was transfered to produce transgenic cotton in an effort to protectec the environment from overuse of pesticides43. It has been reported that microbial chitinases from many fungal strains were stable at broad ranges of temperature and pH, while showcased their effectiveness against graminaceous stem borers Eldana saccharina along withother lepidopteron pests44. Chit1 was isolated from EPF (T. longibrachiatum), which belongs to family 18 of glycosyl hydrolases45. Insoluble chitin can bind specifically with chitinase using the chitin-binding domain46. Endochitinase genes from T. harzianum had been isolated and expressed in potatoes and tomatoes, and it had shown promising results without any adverse effect on the growth and development of both test plants47.
Our study was aimed to evaluate the transient expression of cotton plants and the overall efficiency of Chit1 to control A. gossypii. The proper functioning of the transgenic product was analysed preliminary through the transference of the desired gene in the target cotton plant. Chitinase enzyme assays were performed to find out endochitinase activity, and transgenic plants with T. longibrachiatum (FCBP-EPF-1491) has exhibited chitinases activity of 0.55 U/mL after six days. While, previously, chitinase activity of 0.585 U/mL was recorded after 4 days of incubation48. The highest chitinase activity after 3 to 4 day of fungus production on SDAY(Sabouraud dextrose agar yeast) media was observed. The activity was 6.96 to 46.49 U/mL after 2 h of fungus incubation with 10% colloidal chitin at 37 ºC49. In our study, the chitinase activity of transgenic plants with Chit1 of T. longibrachiatum was recorded as 0.55 U/mL. Chitinase production by T. longibrachiatum was observed50, and maximum production was 6.45 U/mL in plants.Various species of Trichoderma produce enzymes of different concentrations depending upon the substrates. In vitro conditions, various species such as T. gamsii, T. virens, T. longibrachiatum and T. asperellum produced maximum chitinase enzyme of about 0.151 U/mL, 0.107 U/mL, 0.228 U/mL and 0.164 U/mL, respectively. Above all, T. longibrachiatum produced a maximum chitinase of 0.228 U/mL under optimal provided conditions51. Production of chitinase varied according to the substrates, pH and temperature. The different isolates of the same fungal strain can produce chitinases in various concentrations. The activity of chitinase production usually alters by changing the incubation period. But, incubation exceeding six days reduced the amount of chitinase enzyme. The chitinolytic activity of most of fungal strains enhanced at the pH range of 4.0 to 5.252. This study explained the overall production of fungal isolate and difference in production of enzymes in both control and inoculated plants.
Chitinases in the plants are usually involved in the safety of plants against damaging plant pathogens. Most of the induced gene plants showed mild symptoms that confirm the successful transformation of the desired gene in plants. In CLCrV, the plants usually produced light colour veins and downward curling of cotton leaves that confirm the difference between transformed and non-transformed cotton plants. In the case of Virus-induced gene silencing in two model plants that are Arabidopsis and N. benthamiana, severe development of symptoms were displayed when compared to the tobacco rattle virus53,54. Our result documented that transgenic plants with Chit1 of T. longibrachiatum recorded the mortality of nymphs of about 38.75% after 144 h of insect feeding. The mortality of adults of A. gossypii started after 120 h of feeding on transformed cotton plants. The maximum adult mortality of 21.67% was recorded for Chit1 trangenic cotton.
The use of chitinase has revolutionised the agricultural sector as it has altered the use of chemical pesticides. Previously, the chitinase from T. longibrachiatum reported to be widely used to reduce the population of root-knot nematode (Meloidogyne javanica)55. Fungal enzymes and culture filtrate of Metarhizium anisopliae and T. harzianum were toxic to cotton bollworms and mosquitos56,57. The difference in mortality percentage and efficacy of bio-control usually depends on the use of fungal strains and species of insects. In the future, it is highly recommended to use synergistic genes to enhance the overall efficiency of bio-control. It is also recommended to meditate on RNAi (RNA interference) to control the most economical agricultural pests.
Conclusion
In conclusion, fungal chitinases can be widely used to control different agricultural pests, including insects and nematodes, at various morphological states. The cotton plant showed great flexibility for the incorporation of Chit1 genes of T. longibrachiatum. Chit1 showed promising results to control A. gossypii. The formulations of fungal chitinases are less in use as compared to Bt formulations. So, there is a huge need to work on formulations, effect of all possible fungal strains and their effects on insect pests to use as bio-control agents at a vast scale to preserve the environment from the lethal and sublethal effects of pesticides. In future, more chitinase gene can be isolated from different entomopathogenic fungi that can be used to enhance the virulence of transgenic cotton.
Materials and methods
Isolation of the chitinase gene from Trichoderma longibrachiatum
The fungal culture of T. longibrachiatum was obtained from the First Fungal Culture Bank of Pakistan (FCBP), Faculty of Agricultural Sciences, University of the Punjab, Lahore, Pakistan. The chitinase gene was amplified from fungal strains of T. longibrachiatum (FCBP-EPF-1491) that were initially isolated from whitefly35. The genomic DNAwas extracted by using the Cetyl Trimethyl Ammonium Bromide (CTAB) buffer method36. The whitefly DNA was extracted in preheated (65 °C) CTAB buffer. The solution was then mixed with chloroform and centrifuged (16,000 rpm for 4 min) to collect the supernant. The supernatant mixed with isopropanol, and again centrifuged (16,000 rpm for 8 min), followed by washing with ethanol (70%). PCR of samples was performed by using primers; Tri_Chit1_F (5′GCATCTGTGATTTTGCATAC3′) and Tri_Chit1_R (′5′GCCAAGAGACTTGAGGTAAG3′)35. The reaction mixture for PCRwas prepared, that had 18.4 μL of nuclease-free water, 1 μL of 10 mM dNTPs, 0.1 μL of Taq polymerase (5 U μL−1, THERMO FISHER SCIENTIFIC), 0.5 μL of 25 mMMgCl2, 2 μL of 1X tag buffer, 0.5 μL of forward and reverse primers (conc. of 10 pmol in 1 μL) and 2 μL of genomic DNAas a template (50 ng μL−1) in it. Amplification of Chit1 was done by initial denaturation at 95 °C for a minute, repeated 35 cycles for denaturation at 95 °C for a minute as the annealing temperatures were followed by denaturation time. The annealing temperature was 45°Cfor T. longibrachiatum for a minute. The extension was performed at 72 °C for 2 min, and the final extension was at 72 °C for 10 min followed by the final hold at 4 °C.
Cloning in pGEM T-easy vector
The final PCR products were purified from the gel according to the protocol of the Thermo Scientific Genomic purification kit (Catalog number: K0721). The products of PCR were then ligated into cloning vector pGEM T-easy and then it is transformed into the DH5α strain of Escherichia coli (E. coli). After a successful transformation, the plasmid was cloned, isolated, and further confirmed before sequencing through restriction using the restriction enzyme EcoRI.
Sequencing and homology analysis
The plasmid was sent to ETON BIOSCIENCES, San Diego, CA, USA, with both M13 forward and reverse primers for sequencing. Sequences for both forward and reverse strands were amassed with DNA SEQMAN PRO's help (https://www.dnastar.com/t-seqmanpro.aspx). The sequence of the vector was then restricted from both ends. The final sequence was then blasted in NCBI for the nucleotides database.
Characterisation of the chitinase gene
The chitinase gene was characterised before expression in plants. The sequences were blasted in the NCBI database, and their percentage homology and evolutionary relationships were premeditated. For comparison, BLAST analysis37 (NCBI; http://www.ncbi/BLAST) was performed to compare the sequence of Chit1 with releavantchitinases already accessible in databases. The open reading frames (ORFs) for the gene were scrutinised to find out the conserved domain with the help of the Conserved Domain Search Service (CD-Searchhttps://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) tool from NCBI38. The overall signal peptide and molecular weight of isolated chitinase was anticipated with software SIGNAL P 3.039 (http://www.cbs.dtu.dk/services/SignalP).
Expression of chitinase (ORF) in cotton plants
Virus-induced gene silencing vector
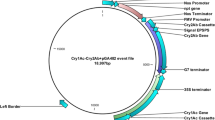
The vector derived from the Cotton leaf crumple virus (CLCrV) was taken from Brown’s lab, University of Arizona, Tucson, AZ, USA. It was a permanent inventory of clones that were already in action to develop cotton plants with transient gene expression40. The ORF coding coat protein of cotton crumple virus (pJRTCLCrVA. 008) was altered with multiple cloning sites (MCS) (Gene bank EU541443) in the Virus-induced gene silencing (VIGS) vector. At the same time, the other component of CLCrV that is B, is also taken from Brown’s lab.
Isolation of chitinase ORF
The open reading frame (ORF) having catalytic domains was isolated from the plasmid. Different primers were designed while keeping information about the restriction sites. Each primer is designed with EcoRI in the forward primer and the NheI restriction site in the reverse primer. ORFs from T. longibrachiatum was amplified from previously cloned chitinases in the pGEM T-easy cloning vector by using Tri_ORF_F (5′ATGAGCTCCTTCGTGCTGGGGTC3′and Tri_ORF_R (5′GAACTAGCATCTGTAATTTTG3′) primers. Employed PCR conditions were as; initial denaturation at 95 °C for 1 min, the second denaturation was at 95 °C for 1 min with repeated 35 cycles followed by annealing temperatures used according to the ORFs for T. longibrachiatum. The annealing temperature was 56 °C for the Chit1 gene of T. longibrachiatum. The time for extension was 2 min at 72 °C with a final extension at 72 oC for 10 min.
Ligation and construction of VIGS-Chit plasmids
The open reading frame of T. longibrachiatum were ligated into the pGEM T-easy vector, and then it was transformed in syrain DH5α of E. coli. Plasmids were isolated, sequenced and analysed before their cloning. VIGS vector (CLCrV-A) and plasmids were restricted with NheI and EcoRI restriction enzymes to clone into the VIGS vector. The ligation was performed at the ratio of 3:1 for both (insert: vector). The restriction analysis was performed for isolation and sequencing of VIGS-Chit recombinant plasmids.
Particle bombardment/ biolistic bombardment
Cultivated cotton seeds (Gossypium hirsutum L.) were collected from the Centre for excellence in Molecular Biology (CEMB), University of the Punjab and placed in pots that had potting soil under control conditions. All methods were carried out in accordance with relevant guidelines. Five seeds were used in each pot, and pots were vigilantly observed until the first true seedlings leaves emerged after 10–15 days. The experiment was divided into five replicates, and each replicate was representing a pot with five seedlings. All of the seedling’s leaves that emerged inoculated biolistically and then were shifted to new individual pots. These pots were placed in a growth chamber with a light period of 15/9 h at 890 μmol m−2 s−1 and temperature of about 23–25 °C. The recommended fertiliser MIRACLE-GRO (Miracle Gro Products, Inc.), was used twice or thrice in a week38 (Anwar et al.). Different temperature ranges as 30 °C/26 °C or 22 °C/18 °C day/night temperatures were set in IAGS, University of the Punjab, with relative humidity (R.H) of 40% to 50% to analyse the growth of seedlings. The plants were individually grown in styrofoam cups with different potting mixtures as 1/3 peat –lite and 2/3 pea gravel and were transferred to 1500 mL pots. Different fertilisers, weak Hoagland solution and water were applied three to four times to all pots. The micro-projectiles made up of gold with 1.5- μm diameter (inBio) coated with a 10 μg mixture of component A having Chit1 ORFs and component B having CLCrV were used for bombardment41. DNA- coated projectiles were loaded with a filter end of Millipore Swinnex and each leaflet was placed below the outlet and bombarded one time from the distance of 6 cm aprroximately. Almost three to four seedlings were bombarded simultaneously with an external pressure of 40–60 psi. For inoculation of seedlings, a commercially available PARTICLE DELIVERY SYSTEM (BioRad PDS1000-He) was used. It has an infection rate of 80%. Both components (A and B) without Chit1 gene were used as mock plants.
Analysis of transgenic expression
The total RNA from transformed cotton plants was extracted by using a QIAGEN kit (catalogue # 74106)41. PCR was performed at 20 °C for 15 min, 37 °C for 120 min, and 10 min at 80 °C followed by 35 cycles. The young cotton leaves were taken after two weeks to confirm the transient expression and their total RNA was extracted. cDNA was produced by the “Revert Aid First Strand cDNA Synthesis Kit” (catalogue #K1622, THERMO SCIENTIFIC). The amplification of the coat protein region of CLCrV was performed using both forward GTTCTAGAATCACCTTCCACTATGAGAC and reverse TCAGAATTCCCTTAACGTGCGATAGATTCTGGGGC primers42. The final PCR products were run on 1.5% agarose gel for the confirmation of transient expression.
Chitinase assay
The activity of recombinant chitinase produced by transgenic cotton was evaluated with the help of the Chitinase Assay kit SIGMA-ALDRICH, catalogue # CS0980. The enzymatic hydrolysis of chitinase substrate was determined. It releases p-nitrophenol upon hydroxylation, and it can be measured at an absorbance of 405 nm. Almost 1.0 μmol of p-nitrophenol was released from one substrate unit per minute at 37 °C and pH 4.8. The substrates used for endochitinase were 4-Nitrophenyl β-D-N, N0, and N00—triacetylchitriose. The purified chitinase was already provided in the kit, and it was used as a positive control.
Virulence bioassay of chitinase transgenic plants against Aphis gossypii
After estimating and confirming various chitinase enzyme activities in cotton plants through chitinase enzyme assays and VIGS vector, transient cotton plants were allowed to feed by 4th instar nymphs and adults of A. gossypii. Data were recorded after regular intervals according to the mortality percentage using modified Abbot’s formula42.
Analysis of data
The final data were analysed and estimated using SPSS, v. 11.5 statistical software (SPSS Inc., Chicago, IL, USA. Before analysis, the percentage data were log transformed.
Data availability
The sequence of chitinase gene of Trichoderma longibrchiatum was submitted to the GenBank (Accession No. OQ689733) in NCBI under the following link: https://www.ncbi.nlm.nih.gov/nuccore/?term=OQ689733 and also provided in related file.
References
Kumar, A. et al. Biocontrol potential of Pseudomonas stutzeri endophyte from Withania somnifera (Ashwagandha) seed extract against pathogenic Fusarium oxysporum and Rhizoctonia solani. Arch. Phytopathol. Plant Prot. 55, 1–18 (2022).
Aksoy, H. M., Saruhan, İ, Kaya, Y. & Ozturk, M. Morphological Changes Caused by Bacillus megaterium on adult emergence of Fall Webworm’s Pupa, Hyphantria cunea (Drury) (Lepidoptera: Erebidae). Tarım Bilim. Derg. 24, 539–546 (2018).
Aslam, M. A., Javed, K., Javed, H., Mukhtar, T. & Bashir, S. Infestation of Helicoverpa armigera Hübner (Noctuidae: Lepidoptera) on soybean cultivars in pothwar region and relationship with physico-morphic characters. Pak. J. Agric. Sci. 56(2), 401–405. https://doi.org/10.21162/pakjas/19.6979 (2019).
Eid, A. E., El-Heneidy, A. H., Hafez, A. A., Shalaby, F. F. & Adly, D. On the control of the cotton aphid, Aphis gossypii Glov. (Hemiptera: Aphididae), on cucumber in greenhouses. Egypt. J. Biol. Pest Control 28, 64 (2018).
Javed, H. Management of eggplant shoot and fruit borer (Leucinodes orbonalis Guenee) by integrating different non-chemical approaches. Pak. J. Agric. Sci. 54, 65–70 (2017).
Malinga, L. N. & Laing, M. D. Efficacy of three biopesticides against cotton pests under field conditions in South Africa. Crop Prot. 145, 105578 (2021).
Hussein, H. S., Tawfeek, M. E. & Eldesouky, S. E. Toxicity and Biochemical Effects of Spirotetramat and its Binary Mixtures with Nanosilica against Aphis gossypii Glover, Bemisia tabaci Gennadius and the Earthworm, Eisenia fetida. Alex. Sci. Exch. J. 43, 107–117 (2022).
Wang, L. et al. Sulfoxaflor resistance in Aphis gossypii: Resistance mechanism, feeding behavior and life history changes. J. Pest Sci. 95, 811–825 (2022).
Chen, T. et al. Detection of stress in cotton (Gossypium hirsutum L.) caused by aphids using leaf level hyperspectral measurements. Sensors 18, 2798 (2018).
Srivastava, R. & Shukla, A. C. Fusarium pallidoroseum: A potential entomopathogenic agent for the biological management of Aphis gossypii. J. Appl. Nat. Sci. 13, 775–785 (2021).
Dubey, J. K. & Sharma, A. Pesticide residues and international regulations. In Sustainable Management of Potato Pests and Diseases (eds Chakrabarti, S. K. et al.) 353–367 (Springer, 2022).
Maurya, R. P. et al. Biological control: A global perspective. Int. J. Trop. Insect Sci. 42, 3203–3220 (2022).
Sayed, S., Elarnaouty, S.-A. & Ali, E. Suitability of five plant species extracts for their compatibility with indigenous Beauveria bassiana against Aphis gossypii Glov. (Hemiptera: Aphididae). Egypt. J. Biol. Pest Control 31, 11 (2021).
Mukhtar, T. Management of Root-Knot Nematode, Meloidogyne incognita, in Tomato with Two Trichoderma Species. Pak. J. Zool. 50, 15 (2018).
Bava, R. et al. Entomopathogenic fungi for pests and predators control in beekeeping. Vet. Sci. 9, 95 (2022).
Yasin, M. et al. Microbial management of ornamental plants/palm common pests. In Microbial Biocontrol: Food Security and Post Harvest Management Vol. 2 (ed. Kumar, A.) 265–284 (Springer International Publishing, 2022).
Gryganskyi, A. P., Golan, J. & Hajek, A. E. Season-long infection of diverse hosts by the entomopathogenic fungus Batkoa major. PLoS ONE 17, e0261912 (2022).
Kidanud, S. Research and application of entomopathogenic fungi as pest management option: A review. J. Environ. Earth Sci. 10, 31 (2020).
Altinok, H., Altinok, M. & Koca, A. Modes of Action of Entomopathogenic Fungi. Curr. Trends nat. Sci. 8, 117–124 (2019).
Aprile, A. M. et al. Combination of the Systemin peptide with the beneficial fungus Trichoderma afroharzianum T22 improves plant defense responses against pests and diseases. J. Plant Interact. 17, 569–579 (2022).
Di Lelio, I. et al. Temperature differentially influences the Capacity of Trichoderma species to induce plant defense responses in tomato against insect pests. Front. Plant Sci. 12, 677830 (2021).
Islam, W. et al. Insect-fungal-interactions: A detailed review on entomopathogenic fungi pathogenicity to combat insect pests. Microb. Pathog. 159, 105122 (2021).
Paschapur, A. et al. Unraveling the importance of metabolites from entomopathogenic fungi in insect pest management. In Microbes for Sustainable lnsect Pest Management: Hydrolytic Enzyme & Secondary Metabolite Vol. 2 (eds Khan, Md. A. & Ahmad, W.) 89–120 (Springer International Publishing, 2021).
Vidhate, R. P., Dawkar, V. V., Punekar, S. A. & Giri, A. P. Genomic determinants of entomopathogenic fungi and their involvement in pathogenesis. Microb. Ecol. 85, 49–60 (2023).
Maharana, C. et al. Secondary metabolites of microbials as potential pesticides. In Sustainable Management of Potato Pests and Diseases (eds Chakrabarti, S. K. et al.) 111–142 (Springer, 2022).
Rajput, M., Kumar, M. & Pareek, N. Myco-chitinases as versatile biocatalysts for translation of coastal residual resources to eco-competent chito-bioactives. Fungal Biol. Rev. 41, 52–69 (2022).
Arakane, Y. & Muthukrishnan, S. Insect chitinase and chitinase-like proteins. Cell. Mol. Life Sci. 67, 201–216 (2010).
Seidl, V., Huemer, B., Seiboth, B. & Kubicek, C. P. A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS J. 272, 5923–5939 (2005).
Graham, W. G. The ecology of chitin decomposition. Adv. Microb. Ecol. 11, 387–430 (1990).
Mathur, N. K. & Narang, C. K. Chitin and chitosan, versatile polysaccharides from marine animals. J Chem. Educ. 67, 938. https://doi.org/10.1021/ed067p938 (1990).
Soesanto, L., Fatihah, B., Manan, A., Mugiastuti, E. & Prihatiningsih, N. Organic control of Bemisia tabaci Genn on Capsicum annuum with entomopathogenic fungi raw secondary metabolites. Biodivers. J. Biol. Divers. 21, d211240 (2020).
Elbanhawy, A. A., Elsherbiny, E. A., Abd El-Mageed, A. E. & Abdel-Fattah, G. M. Potential of fungal metabolites as a biocontrol agent against cotton aphid, Aphis gossypii Glover and the possible mechanisms of action. Pestic. Biochem. Physiol. 159, 34–40 (2019).
Ozyigit, I. I., Dogan, I., Kaya, Y., Bajrovic, K. & Gozukirmizi, N. Cotton biotechnology: An efficient gene transfer protocol via Agrobacterium tumefaciens for a greater transgenic recovery. J. Nat. Fibers 19, 11582–11596 (2022).
Hoell, I. A., Klemsdal, S. S., Vaaje-Kolstad, G., Horn, S. J. & Eijsink, V. G. H. Overexpression and characterization of a novel chitinase from Trichoderma atroviride strain P1. Biochim. Biophys. Acta BBA 1748, 180–190 (2005).
Anwar, W. et al. First record of Trichoderma longibrachiatum as entomopathogenic fungi against Bemisia tabaci in pakistan. Pak. J. Phytopathol. 28, 287–294 (2016).
Gontia-Mishra, I., Tripathi, N. & Tiwari, S. A simple and rapid DNA extraction protocol for filamentous fungi efficient for molecular studies. (2014).
Stenglein, S. A. & Balatti, P. A. Genetic diversity of Phaeoisariopsis griseola in Argentina as revealed by pathogenic and molecular markers. Physiol. Mol. Plant Pathol. 68, 158–167 (2006).
Anwar, W. et al. Chitinase genes from Metarhizium anisopliae for the control of whitefly in cotton. R. Soc. Open Sci. 6, 190412 (2019).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Idris, A. M., Brown, J. K. & Robertson, D. Geminivirus-mediated gene silencing from cotton leaf crumple virus is enhanced by low temperature in cotton geminivirus-mediated gene silencing from cotton leaf crumple virus is enhanced by low temperature. Plant Physiol. https://doi.org/10.1104/pp.108.123869 (2008).
Kjemtrup, S. et al. Gene silencing from plant DNA carried by a Geminivirus. Plant J. 14, 91–100 (1998).
Fleming, R. & Retnakaran, A. Evaluating single treatment data using abbott’s formula with reference to insecticides. J. Econ. Entomol. 78, 1179–1181 (1985).
Moreira, M. F. et al. A chitin-like component in Aedes aegypti eggshells, eggs and ovaries. Insect Biochem. Mol. Biol. 37, 1249–1261 (2007).
Okongo, R. N., Puri, A. K., Wang, Z., Singh, S. & Permaul, K. Comparative biocontrol ability of chitinases from bacteria and recombinant chitinases from the thermophilic fungus Thermomyces lanuginosus. J. Biosci. Bioeng. 127, 663–671 (2019).
Gentile, A. et al. Enhanced resistance to Phoma tracheiphila and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed. 126, 146–151 (2007).
Blaak, H. & Schrempf, H. Binding and substrate specificities of a Streptomyces olivaceoviridis chitinase in comparison with its proteolytically processed form. Eur. J. Biochem. 229, 132–139 (1995).
Cumagun, C. J. R. Managing plant diseases and promoting sustainability and productivity with Trichoderma: The Philippine experience. J. Agric. Sci. Technol. B 14, 699–714 (2012).
Zhang, S., Gan, Y., Xu, B. & Xue, Y. The parasitic and lethal effects of Trichoderma longibrachiatum against Heterodera avenae. Biol. Control 72, 1–8 (2014).
Mustafa, U. & Kaur, G. Studies on extracellular enzyme production in Beauveria bassiana isolates. Int. J. Biotechnol. Amp Biochem. 6, 701–714 (2010).
Kovacs, K., Szakacs, G., Pusztahelyi, T. & Pandey, A. Production of chitinolytic enzymes with Trichoderma longibrachiatum IMI 92027 in solid substrate fermentation. Appl. Biochem. Biotechnol. 118, 189–204 (2004).
Osorio-HernÃndez, E. et al. In vitro activities of Trichoderma species against Phytophthora parasitica and Fusarium oxysporum. Afr. J. Microbiol. Res. 10, 521–527 (2016).
Mishra, S., Kumar, P. & Malik, A. Effect of process parameters on the enzyme activity of a novel Beauveria bassiana isolate. Int. J. Curr. Microbiol. Appl. Sci. 2, 49–56 (2013).
Ratcliff, F., Martin-Hernandez, A. M. & Baulcombe, D. C. Technical Advance: Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245 (2001).
Turnage, M. A., Muangsan, N., Peele, C. G. & Robertson, D. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30, 107–114 (2002).
AL-Shammari, T. A., Bahkali, A. H., Elgorban, A. M., ElKahky, M. T. & Al-Sum, B. A. The use of Trichoderma longibrachiatum and Mortierella alpina against Root-Knot Nematode, Meloidogyne javanica on Tomato. J. Pure Appl. Microbiol. 7, 199–207 (2013).
Mohanty, S. S. & Prakash, S. Effects of culture media on larvicidal property of secondary metabolites of mosquito pathogenic fungus Chrysosporium lobatum (Moniliales: Moniliaceae). Acta Trop. 109, 50–54 (2009).
Binod, P., Sukumaran, R. K., Shirke, S. V., Rajput, J. C. & Pandey, A. Evaluation of fungal culture filtrate containing chitinase as a biocontrol agent against Helicoverpa armigera. J. Appl. Microbiol. 103, 1845–1852 (2007).
Acknowledgements
This work was supported by Faculty of Agricultural Sciences and First Fungal Culture Bank of Pakistan (FCBP), University of the Punjab.
Author information
Authors and Affiliations
Contributions
K.A.Z., W.A., H.A., T.A. contributed to the study conception and design. Material preparation, data collection and analysis were performed by W.A., A.A., U.B. and M.A.J. The first draft of the manuscript was written by R.K. and H.A.A.K. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Anwar, W., Amin, H., Khan, H.A.A. et al. Chitinase of Trichoderma longibrachiatum for control of Aphis gossypii in cotton plants. Sci Rep 13, 13181 (2023). https://doi.org/10.1038/s41598-023-39965-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-39965-y
This article is cited by
-
Identification of mycoparasitism-related genes in Trichoderma harzianum T4 that are active against Colletotrichum musae
Archives of Microbiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.