Abstract
In this work, an analytical model is developed for DM-DG-TMD-FET- based Biosensor including Fringing-field effects. The Analytical model has been developed for two different Device structures, namely Device structure-1 (without a gate above the nano-cavity) and Device structure-2 (with a gate above the nano-cavity) based on modulation of the dielectric constant of biomolecules in the nano-cavity region. The proposed model has been validated against both numerical quantum simulation results with the help of a few fitting parameters and it also agrees with the 2-dimensional numeric simulator SILVACO TCAD used in this work. The presence/absence of biomolecules has been detected by the metric of threshold voltage sensitivity \(S_{Vth}\) and drain current \(I_{d}\) for the neutral as well as charged biomolecules. Sensitivities of partially filled nano-cavities arising out of steric hindrance in both the biosensors are compared. Optimization of device dimensions has also been included in this work to enhance the sensitivity of the biosensors. It has been witnessed that the sensitivity of the proposed biosensor is \(\sim\) 100% higher in Device structure-1 for neutral biomolecules with dielectric constant \(\kappa\) = 12, when compared to Device structure-2 for fully filled cavities. Whereas for the charged biomolecules, Device structure-1 shows \(\sim\) 50% enhanced sensitivity than Device structure-2 for \(N_{f}=-1\times 10^{-12}\) \(\text{C}/\text{cm}^2\). Device structure-1 demonstrates \(\sim\)120% higher sensitivity than Device structure-2 with partially filled cavities (i.e. 66% filled cavity). Finally, benchmarking of the proposed biosensor is presented with contemporary, state-of-the-art biosensors and it is highlighted that \(MoS_{2}\) FET-based biosensor emerges with a superior sensitivity of \(S_{Vth}\) = 0.81 V for \(\kappa = 12\).
Similar content being viewed by others
Introduction
FET-based biosensors have been extensively investigated to detect a range of target biomolecules due to their superior sensitivity, swift label-free detection capabilities, low power consumption, compatibility with CMOS technology, and large scale production at a low cost. These factors have become more critical as the demand for ultrasensitive biosensors has risen in fields such as medicine, agriculture, defence, and environmental monitoring, among others1,2,3. Among FET-based biosensors, Dielectric Modulated (DM) FETs have shown better sensitivity and are mostly preferred in the field of biosensing applications as the created nano-cavities provide sufficient volume and efficient conjugation of biomolecules which results in enhancement in sensitivity4. Choi et al. demonstrated the first DM-FET-based biosensor with nano-cavities for label-free detection of biomolecules2. After that many dielectric-modulated FET-based biosensors have been reported with the enhancement in sensitivity5,6,7,8,9.
FET-based biosensors with 1-D Nanowires and CNTs as channel material have a significant advantage over label-based detection10 because they exhibit good electrostatics between biomolecules and the channel, resulting in increased sensitivity. However, synthesizing biosensors that meet all of these criteria at the same time has proven difficult, as 0-D and 1-D nanomaterials-based biosensors are difficult to fabricate on a large scale and have high production costs11,12,13, limiting the practical feasibility of such structures.
Contrarily, tremendous progress has recently been made with Transition Metal Dichalcogenides (TMDs), one of the most intriguing 2-D semiconducting materials for next-generation biosensors. Because of their excellent electrostatic control, planar nature, high surface-to-volume ratio, superior charge sensitivity, and high electron mobility, among other advantage, TMDs hold a lot of promise for use as a channel in FET-based ultra-sensitive biosensors12,14 .
Furthermore, due to their comparably weak interlayer connections, 2-D materials have a lower surface roughness than 3-D materials15. TMD materials unlike Graphene exhibit a band-gap that is essential for the operation of FET-based biosensors since the binding process at the interface between the channel and biomolecules modulates the carrier transport in 2-D layered materials16.
Moreover, in recent years the application of various 2-D layered materials like \(MoS_{2}\), \(WSe_{2}\) and \(MoSe_{2}\) in FET-based biosensors has become extremely promising. \(MoSe_{2}\) has smaller band-gap than \(MoS_{2}\) and \(WSe_{2}\) but it exhibits larger reduced effective mass which is preferable for Tunnel FET-based applications17,18. Whereas, \(WSe_{2}\)-based biosensors exhibit higher linear-regime sensitivities in comparison with \(MoS_{2}\)-based biosensors19. Because of the recent advances in the synthesis of \(MoS_{2}\) sheets using CVD and liquid phase exfoliation techniques12,16, as well as their excellent compatibility with commercial planar processes for large scale production15,20, \(MoS_{2}\) FET-based biosensors are widely investigated among TMD materials.
Sarkar et al. have reported detection of streptadavin using \(MoS_{2}\) FET-based biosensor with \(HfO_{2}\) as gate dielectric functionalized with biotin11. Wang et al. have reported APTES functionalized \(MoS_{2}\) nanosheet-based biosensor for the cancer marker protein detection21. Nam et al. have reported \(MoS_{2}\) FET-based biosensor functionalized with \(TNF {\text{-}}\alpha\) anti-bodies for sensing of \(TNF{\text{-}}\alpha\) molecules22.
However, in order to further investigate 2-D material based FET in biosensing applications, an analytical I-V model that accounts for realistic circumstances when conjugating the biomolecules in the nano-cavities, which seem to be unavoidable during fabrication, is required. Unfortunately, little progress has been made in developing various analytical models for DM-TMD-FET-based biosensors. An I-V model has been developed by Rahman et al.23 for single-sided cavity, \(MoS{_2}\)-based biosensor. However, this model does not take into account fringing-field effects, which in sub-100 nm FETs cannot be disregarded. Furthermore, the author has investigated the effect on sensitivities when the nano-cavities are filled completely, which is practically impossible.
Main contributions of this work can be summarized as: an analytical model of DM-TMD-FET-based biosensor has been developed including several design parameters like nano-cavity thickness, dielectric constant and charge of biomolecules. The veracity of the model has been verified with experimental data24 and 2-D numeric simulator SILVACO TCAD25. Two device structures namely, Device structure-1 (without gate above the nano-cavity) and Device structure-2 (with gate above the nano-cavity) have been investigated incorporating fringing-field effects in the model and its effect on the characteristics of the biosensors has been analyzed. Sensitivity metrics such as threshold voltage \(V_{th}\), shift in threshold voltage \(\Delta\) \(V_{th}\) and drain current are estimated for neutral biomolecules as well as charged biomolecules. Partially filled nano-cavities with different filling factors have also been considered in this work and the analytical model has been developed to incorporate this. The effect of steric hindrance including concave, convex, decreasing, and increasing step profiles in the nano-cavity has been considered and the impact of these profiles on the sensitivity has been observed.
Device structure
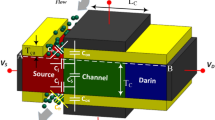
Figure 1a,b show the schematic of the proposed monolayer TMD-FET-based biosensor without gate over cavity and with gate over cavity.
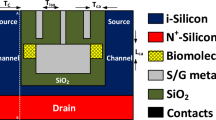
An equivalent capacitance model of the device structure-1 and 2 has also been drawn in Fig. 2. Here, x is the channel direction and, y is perpendicular to the channel. The channel region is divided into three parts, regions at both the source (\(L_{1}\)) and the drain side (\(L_{3}\)) work as cavity, where biomolecules can be immobilized, region beneath the high-\(\kappa\) gate oxide is the overlap region, (\(L_{2}\)). A \(SiO_{2}\) layer having thickness 1 nm is taken in the cavity regions, as illustrated in Fig. 1, which acts as an adhesion layer for the biomolecules3. Since, \(MoS_{2}\)-based biosensor has been widely explored26,27 in the recent years, it has been utilized as a channel material for performing various analysis in this work. Other TMD materials like, \(WSe_{2}\) and \(MoSe_{2}\) can also be deployed as channel materials28. TMD-FET-based biosensor under consideration consists of a monolayer p-doped \(MoS_{2}\) channel sandwiched between the top and the bottom \(HfO_{2}\) layer. Al is used as top and bottom gates. Highly n-doped monolayer \(MoS_{2}\) has been used in the source and the drain regions. The channel has a thickness and length of 0.7 nm19 and 60 nm respectively. On the source and drain sides, the cavity is 20 nm long, respectively. The top and bottom oxide layers are each 10 nm thick. The remaining model parameters are listed in Table 1. A tentative fabrication process20,27,29 of the proposed TMD FET-based biosensor has been depicted in Fig. 3.
Results and discussions
The characteristics of the DM-DG-TMD-FET-based biosensor for neutral and charged biomolecules are discussed in this section. The sensitivity of the biosensor is measured in terms of threshold voltage (\(V_{th}\)), variation in threshold voltage (\(\Delta V_{th}\)) and drain current (\(I_{ds}\)).
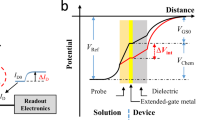
The proposed model is validated with the experimental I-V characteristics of monolayer \(WSe_{2}\) by Fang et al.24, quantum simulation results obtained by Rahman et. al.23 and with two dimensional numerical simulator Silvaco TCAD25. Exactly same sets of the device and material parameters are utilized to validate our proposed model with the simulator. Figure 4 depicts the I-V characteristics of the DM-DG-TMD- FET- based biosensor obtained from the model along with the experimental results reported by Fang et al.24. Figure 5 shows the surface potential of DM-DG-TMD FET-based biosensor, which has been validated with self consistent NEGF simulation results of23. Drain current and surface potential shows a close match with the results from24 and23. Little discrepancy exists as a result of the various environmental set-up.
Drain current, \((I_{ds})\) versus Gate to source bias, \((V_{gs})\)) for back-gate, (Vgb) − 40 V and drain to source bias, \(V_{ds}\)= − 0.05 V with channel-length, \(L_{ch}\) = 9.4 nm, front and back gate oxides are \(ZrO_{2}\) \(T_{tox}=17.5\; \text{nm}\), \(k_{tox}=12.5\) and \(SiO_{2}\) \((T_{box}=270 \; \text{nm}, k_{box}= 3.9)\) respectively.
Surface potential calculated using our proposed model vs. quantum simulated data of23 for \(V_{gs}=1 \; \text{V}\).
Impact of neutral biomolecules and charged biomolecules on sensitivity
In this section, we have explored the impact of neutral biomolecules and charged biomolecules on the surface potential, threshold voltage \(V_{th}\) and drain current of the proposed two structures of DM-DG-TMD-FET-based biosensor. Figure 6a shows the bending in surface potential when neutral biomolecules get immobilized in the cavity for the proposed DM-DG-TMD- FET-based biosensor, structure-1. In Device structure-1, there is no gate over nano-cavity so the effect of flat band voltage \(V_{fb}\) is negligible and fringing capacitance plays a dominant role by influencing the surface potential. Deviation in the surface potential profile is clearly observed under the nano-cavity region, whereas, no deformation of the potential profile is observed in the region without a nano-cavity.
When the cavity is empty i.e filled with air (\(\kappa =1\)) the gate capacitance is low and the flat band voltage \(V_{fb}\) in the cavity part is negligible so the voltage requirement will be more for the electrons to cross the source-channel barrier which is evident from (1). The \(V_{th}\) of a MOSFET can be expressed as30
Moreover, when the dielectric constant of the biomolecules increases, the effective gate capacitance rises along with fringing-field capacitance, changing the surface potential in the cavity region. Thus with an increase in the dielectric constant of biomolecules the fringing-field effects dominates and the surface potential barrier is reduced.
For the proposed Device structure-2, Fig. 6b illustrates the deformation in surface potential when neutral biomolecules get immobilised in the cavity. Since there is gate over cavity, fringing capacitance is not dominant in this case and the control of gate over channel is adequate, as the channel is p-type, there will be negative flat band voltage across the channel and the amount of threshold voltage for the electrons to cross the source channel barrier will be less. In this case also the surface potential barrier decreases with the increase in dielectric constants of the biomolecules but the reduction in barrier is more when compared to Device structure-1 because of the presence of gate control throughout the channel region and sufficient amount of flat band voltage \(V_{fb}\).
When the target biomolecules are immobilized in the nano-cavities, device characteristics are modulated according to the quantity of target biomolecules. The threshold voltage \(V_{th}\) and change in threshold voltage \(S_{Vth}\) are the most commonly utilised sensing metrics for evaluating the effectiveness of the biosensors among the different parameters that are affected by the target biomolecules.
It is evident from Fig. 7a,b, that the threshold voltage requirement for Device structure-1 is higher than that for Device structure-2, as the gate control over the channel in structure-1 is less so the \(V_{fb}\) is negligible in the cavity area and threshold voltage increases and due to the fringing effect there is more increase in threshold voltage. For Device structure-2 there is full gate control over the channel thus threshold voltage requirement is less. It is also evident from Fig. 7a,b, that the threshold voltage decreases with the increasing \(\kappa\). When the cavity is empty (i.e \(\kappa =1\)), the threshold voltage achieved is high, i.e 0.969 V for structure-1 and and 0.569 V for structure-2.
Figure 7a,b plots the sensitivity of the proposed two structures of DM-DG-TMD-FET-based biosensors. The sensitivity of the biosensor is measured by a shift in threshold voltage (\(S_{Vth}\)) before and after immobilization of the biomolecules in the nano-cavity region. Thereby, it can be defined as31
It can be observed that sensitivity of device structure-1 is \(\sim 100\%\) more than the sensitivity of device structure-2. As in the proposed biosensor structure-1, fringing capacitance dominates the total capacitance which leads to increase the threshold voltage as well as overall enhancement in the sensitivity.
Figure 8a shows the drain current variation when neutral biomolecules are conjugated in the nano-cavity region of the proposed Device structure-1. The OFF current of the device is minimum when the cavity is empty and it increases when the dielectric constant \(\kappa\) in the cavity region increases, whereas, there is slightly increment in ON current. Now, comparing Device structure-2 Fig. 8b to Device structure-1, the ON and OFF current increases as the dielectric constants \(\kappa\) increase. The variation in OFF current is greater than the variation in ON current because FETs conduct extremely low current in the subthreshold regime, so a smaller change in potential due to biomolecule immobilization leads to a reasonable change in subthreshold current, which is preferable for sensing operation.
Figure 9a,b shows the effect of charged biomolecules on the threshold voltage of the proposed structure. The impact of charged biomolecules on the sensitivity of the device is also depicted in Fig. 9a and b for both the proposed structures. The sensitivity parameter \(S_{Vth}\) for negatively charged biomolecules in the figure is considerably higher than that for positively charged biomolecules. Additionally, the sensing metric rises in accordance with an increase in the charge density of negatively charged biomolecules. Sensitivity, on the other hand, first show a linear trend with an increase in charge density after that it get saturated to further variations. Before and after immobilising charged molecules, the proposed device’s sensitivity can be described as,
Since, there is gate over the nano-cavity (Device structure-2) regions, thus, the threshold voltage requirement to deplete the channel will reduce by the amount of \(V_{fb}\) compared to Device structure-1 for negatively/positively charged biomolecules, which can be understood from (20). For negatively charged biomolecules the threshold voltage requirement to deplete the channel is more and it is even more for device structure-1 because of the presence of fringing field effects. Which leads to decrement in ON and OFF current for negatively charged biomolecules. Whereas, for positively charged biomolecules, the threshold voltage requirement to deplete the channel is less for both device structures and it is even lesser for Device structure-2 because of the absence of fringing effects. Which leads to increment in ON and OFF current as shown in Fig. 10.
Sensitivity analysis with the variation in thickness and length of the nanogap cavity
In this section we have demonstrated how the device’s sensitivity varies with the thickness \(T_{cav}\) and length \(L_{cav}\) of the cavity. Figure 11a shows the change in threshold voltage \(S_{Vth}\) with the variation in thickness of the cavity from 7 to 9 nm. With the increase in thickness of the cavity, barrier between source/channel junction increases, results in decrement of the drain current so, as expected \(V_{th}\) linearly increases as \(T_{cav}\) becomes thicker. The variation in sensitivity parameter with cavity length \(L_{cav}\) varying from 30 to 40 nm of channel is shown in Fig. 11b. As the cavity length \(L_{cav}\) increases, requirement of threshold voltage increases which leads to low drain current and higher sensitivity, which further improves if the channel length is large as also reported for DM-FET6.
Impact of partially filled biomolecules on sensitivity
All of the preceding sections assumed that the cavities were completely filled with biomolecules, but in reality, entirely filled cavities are rarely possible.
Thus, three distinct assumptions of biomolecules filling the nano-cavity are investigated here, as illustrated in Fig. 12. The fill-factor of the nano-cavity is defined as the ratio of the area covered by conjugated biomolecules to the overall area of the nano-cavity in percentage. This section discusses the effect of percentage volume filling of the nano-cavity on the sensitivity of DM-DG-TMD-FET-based biosensor. The fill-factor (\(\rho _{T_{bio}}\)) defined here as,
Fill-factor of the completely filled cavity is 100%. For the simulation purpose, \(L_{1}\) and \(L_{2}\) are fixed at 35 nm and \(T_{bio}\) is taken to be 6 nm for 66% filled cavity and 4 nm for 50% filled cavity and 2 nm for 33% filled cavity. The effect of partially filled cavities on the surface potential can be understood through (47) and (48). For different percentage filling of profiles (47) is fed to (24) and (25) for Device structure-1 and for Device structure-2 (48) is used and surface potential for partial filled cavity is calculated using (32) and (33). Figure 13 depicts the surface potential for various assumptions of a filled cavity. The figure shows that as \(T_{bio}\) is reduced for non-fully filled cavities, the number of biomolecules inside the cavities decreases and the effective gate capacitance decreases, resulting in less gate control. As a result, the source-to-channel barrier increases. Figure 14 shows the sensitivity for different dielectric constants of biomolecules for the fill in factor of 66%, 50% and 33%.
Sensitivity is observed to enhance with increase in fill-factor from 33% to 100% at a specific value of \(\kappa\). The maximum sensitivity is shown for a fully filled cavity since the amount of biomolecules conjugation increases as the fill-factor increases. Additionally, it is observed that sensitivity rises as both the fill-factor and the value of \(\kappa\) increase.
Sensitivity analysis due to existence of steric hindrance
Biomolecules are immobilized and hybridized inside the cavity for biosensing operations. Existing hybridized biomolecules block the admission of fresh biomolecules during the process of biomolecule hybridization.
This steric hindrance effect may cause biomolecules to hybridize in non-uniform ways along the length of the cavities, according to7. Thus, in practical-scenario, completely filled cavities are not conceivable. In this work four different step profiles viz. decreasing, increasing, concave, and convex as shown in Fig. 15a–d to depict the issue of steric hindrance associated with arbitrary and random biomolecular profiles has been considered. A comparison of sensitivities among these profiles has been presented with \(\sim\)100% filling factor in the cavitites. For decreasing step profile, shown in Fig. 15a, biomolecules \(\kappa >1\) will gather nearer to the source/channel junction7, and thus coupling between gate and channel enhances. As a consequence, the tunneling rate will speed up and measurable variation on \(V_{th}\) happens. Hence, this provide improvement in sensitivity. For the rest of the profiles biomolecules are away from the source to channel interface which reduce the gate to channel coupling and offer poor sensitivities. Decreasing profile of both the structures shows maximum sensitivity. Decreasing profile of Device structure-1 shows sensitivity \(S_{Vth}\) of 0.789 V while decreasing profiles of Device structure-2 shows 0.28 V of sensitivity as depicted in Fig. 16a,b.
Benchmarking
A large number of biosensors based on analytical modelling and simulation are reported in literature. Here, the sensitivity of monolayer DM-DG-TMD-FET-based biosensor is compared with other reported alternative FET-based biosensors. The estimated sensitivities of FET-based biosensors are extracted from literature and compared with this work. Comparative magnitude of sensitivity metrics form different literature is depicted in Table 2. It is observed from Table 2 that the sensitivity of the proposed Device structure-1 with \(MoS_2\) as channel material shows highest sensitivity among all the FET-based biosensor structures considered for benchmarking purpose. So, it is envisaged that \(MoS_2\)-based proposed Device structure-1 can be a potential candidate for biosensing applications with extremely high sensitivity.
Modelling and simulation methods
In40, it is shown that 1-D Poisson’s equation can be a rational approximation to explain the electrostatic behavior in 2–D TMD FETs. Surface potential modelling in this work is also based on the solution of the 1-D Poisson’s equation in the channel40. Surface potential distribution \(\phi (x)\) in the 2–D channel region can be expressed as,
Where \(\varepsilon _{tox}\) and \(\varepsilon _{box}\) are top and bottom gate dielectric constants, respectively. Here \(T_{tox}\) and \(T_{box}\) are top and bottom gate oxide thicknesses respectively, \(\varepsilon _{ch}\) is the dielectric constant of 2-D material-based channel. \(N_{A}\) is the acceptor type dopant concentration per unit area and \(n_{2D}(x)\) is the free inversion carrier concentration. \(V'_{Gt}\) and \(V'_{Gb}\) are top and bottom gate voltages, respectively defined as,
\(V_{Fbt}\) and \(V_{Fbb}\) are flat band voltages of top and bottom gate, respectively. where,
Where \(N_{dos}\) is the effective density of states of 2-D channel.
Differentiating (5) with respect to x and substituting the value of \(\frac{q}{\varepsilon _{ch}T_{ch}} (n_{2D}(x))\) from (5) into the result, we obtain,
The differential equation in (11) cannot be expressed in a closed form analytical solution. Gradual channel approximation is invoked to simplify (11) and equated as \(\frac{\delta V}{\delta x}=0\)41. (11) can be simplified further by ignoring the variations of higher order \(\Phi\) (x) with x. As long as the channel is long and drain bias is low, this assumption is valid and it be can expressed as,
As, lateral electric field is non-zero i.e. \(\frac{\delta V}{\delta x}\ne 0\), so, when voltage is applied to the drain, (12) reduces to a linear differential equation as,
Where,
The closed form solution of the differential equation in (12) can be expressed as,
Modelling of surface potential in overlap region (region II)
In the gate overlap channel region i.e. region II (Fig. 1) Surface potential distribution can be expressed as,
Following boundary conditions need to be satisfied in this region for the continuity of potential and electric displacement at the interfaces,
Modelling of surface potential in nano-cavity region (region I and region III)
In this subsection, detailed modelling of surface potential in the nano-cavity region (Fig. 1) is discussed. Flatband voltage expressions in the nano-cavity regions L1 and L3 regions in Fig. 1) can be written as,
\(V_{fb1}\) and \(V_{fb3}\) are the flat band voltages of region I and region III, respectively. Here \(C_{fr}\) is the gate electrode fringing capacitance. Fringing-field is modeled using the conformal mapping technique, the mapping functions can be expressed as42,43,44,
The structure after transformation of the device and the orgin of fringing field is shown in Fig. 17. Now using structural transformation technique42,43,44, the fringing capacitance can be expressed as,
Cross-section of the Device structure 1 (Fig. 1a) showing the fringing field lines and the structural transformation.
Modelling of surface potential in nano-cavity region (region I and region III) for Device structure-1
For the proposed DM-DG-TMD-FET structure-1 mentioned in (Fig. 1a), when the fringing fields are dominant, the effective capacitance is determined primarily by the fringing capacitance.
So, for the region I (\(L_{1}\)) of the Device structure-1 shown in (Fig. 1a), the closed form expression of surface potential can be written as
For region III (\(L_{3}\)), the closed form expression of surface potential can be written as,
Where,
Modelling of surface potential in nano-cavity region (region I and region III) for Device structure-2
For region I (\(L_{1}\)) of the Device structure-2 in (Fig. 1b), the closed form expression of surface potential can be written as,
For region III (\(L_{3}\)), the closed form expression of surface potential can be written as,
Where,
For the proposed DM-DG-TMD-FET structure-2 mentioned in (Fig. 1b), when the fringing fields are less dominant, the effective capacitance is determined primarily by the gate capacitance.
Here, \(C_{gap}\) is the capacitance of cavity region and it can be expressed as,(\(\frac{\varepsilon _{bio}}{T_{cav}}\)). For finding out the constant coefficients, \(C_{1}\), \(C_{2}\), \(C_{3}\), \(C_{4}\), \(C_{5}\) and, \(C_{6}\), following boundary conditions are enforced.
Solving (42), (43), (44) and, (45), the constant coefficients can be evaluated and then substituting the values of constants in (24), (25), (32) and (33) surface potential can be evaluated.
Surface potential modelling for partially filled cavity
In this section, surface potential is modeled for the mentioned device structures in (Fig. 1) with partially filled nano-cavity which is filled upto a certain height, \(T_{bio}\). For Device structure-1 (Fig. 1a) \(T_{cav}\)=\(T_{bio}\) considered as 10 nm and for Device structure-2 (Fig. 1b) as 9 nm (this is for fully filled case). For partially field case, \(T_{bio}\) can be treated as a parameter. Gate electrode fringing capacitance when the nano-cavity of region I and region III is partially filled upto a certain height (\(T_{bio}\)) can be expressed as,
Where
where \(T_{bio}\) is the height of biomolecules occupancy and \(\varepsilon _{bio}\) is the dielectric constant of biomolecules. By substituting (47) in (24) and in (25) surface potential for partially cavity case can be obtained. For the proposed Device structure-2, effect of (48) will get added to (40) and from (32) and (33), surface potential for partially filled cavity case can be obtained.
Drain current modelling
Model of drain current in subthreshold region is calculated by utilizing expressions of surface potential obtained in previous section45 and can be expressed as,
For linear regime, three transistor modelling approach has been employed to calculate the drain current46. Both the subthreshold current and linear regime current have been equated at transition region for the continuity of the current. The carrier transport is governed by the drift-diffusion equation described as,
Where \(\mu (x)\) is carrier mobility in the channel, and W is device width. Here an extraction of V(x) in terms of x to calculate the drain current is required. We assume a linear profile of V(x) and simplified expression of potential can be written as
The constants M and C can be evaluated as mentioned in23. To incorporate the effect of gate bias, an empirical fitting function \(F(V_{G})\) can be considered with C. So, the final form of C can be expressed as,
Now integrating (50) w.r.t to x, current in each region can be evaluated. The drain current in region I,II,III (Fig. 1) is the drain current of the DM-DG-TMD-FET with channel length, \(L_{1}\), \(L_{2}\), and \(L_{3}\) respectively, and it can be expressed as,
Where, \(i= 1,2,3\).
Simulation methodology
The developed model of the TMD FET-based biosensor has been validated with the 2-dimensional numeric simulator SILVACO TCAD25. Atomistic simulators such as Nano TCAD ViDES is compatible for atomistics simulations of TMD devices but they suffer from computational complexity and it has its own limitations. Moreover, many recent work based 2-D materials FET have been using SILVACO TCAD for simulating FETs18,47. In SILVACO TCAD, while defining 2-D materials, the electrical properties of materials like permittivity, densitiy of states, electron and hole effective masses, affinity, electron and hole mobilities need to be defined. For simulation of the proposed device we have adopted the fermi-dirac carrier distribution, the electric field-dependent mobility (FLDMOB) model concentration dependent mobilty (CONMOB) model. Shockley- Read-Hall recombination model is also included for the recombination mechanism. As the source and drain regions are highly doped regions, a significant amount of band bending is there in these regions due to high doping, to include this in calculations, Band-gap narrowing model(BGN) has also been considered. The effect of neutral biomolecules is modeled by introducing a dielectric materials having dielectric constant (\(\kappa\)) varied between 1 to 12, which lies in the range of dielectric constant of different biomolecules (e.g., Protein (\(\kappa\) = 2.50), Biotin (\(\kappa\) = 2.63), APTES (\(\kappa\) = 3.57), Protein (\(\kappa\) = 6) and Streptavidin (\(\kappa\) = 2.1)5,48). Negative and positive charge density (\(N_{f} = -1\times 10^{-12}\) \(\text{C}/\text{m}^2\) to \(+1\times 10^{11}\) \(\text{C}/\text{m}^2\)) at the \(MoS_{2}/SiO_{2}\) interface is considered to model the impact of charged biomolecules. Material parameters of the TMD materials have been extracted from47.
Conclusion
In this work, for monolayer \(MoS_{2}\)-based dual-gate FET-based biosensor, an analytical model has been developed including the fringing-field effect. The model is developed in such a way that it can be utilized to design DM-DG-TMD FET-based biosensor based on the principle of modulation of dielectric constants. The proposed model can be applied to any TMD material to measure its impact on sensitivity. The proposed biosensor shows \(\sim 100\%\) higher sensitivity in Device structure-1 compared to Device structure-2. Channel surface potential and transfer characteristics obtained from the model show a quantifiable variation in sensor output with the variation of dielectric constant in the nano-cavity region. The effect of partially filled cavities has been also modeled and investigated in this work. Steric hindrance issue has also been studied in this work. The results demonstrate that the completely filled cavity shows \(\sim 92\%\) higher sensitivity than the partially filled cavities (i.e. 33% filled cavity). Optimization of device dimensions has also been done to enhance the sensitivity of the proposed biosensors. Finally, benchmarking of the proposed biosensor is presented with contemporary, state of the art biosensors and it is highlighted that \(MoS_{2}\) FET-based biosensor emerges with a superior sensitivity \(S_{Vth}\) = 0.81 V for \(\kappa =12\). Finally, it is concluded that our proposed DM-DG-TMD FET-based Biosensor structure-1 model is an extremely promising candidate for the biosensing applications due to its enhanced sensitivity and label-free detection trait which could be immensely useful for detection of lower concentration of biomolecules.
Data availability
Data can be available upon reasonable request to the corresponding author.
References
Gu, B. et al. Nanogap field-effect transistor biosensors for electrical detection of avian influenza. Small (Weinheim an der Bergstrasse, Germany) 5(21), 2407–2412. https://doi.org/10.1002/smll.200900450 (2009).
Im, H., Huang, X. J., Gu, B. & Choi, Y. K. A dielectric-modulated field-effect transistor for biosensing. Nat. Nanotechnol. 2(7), 430–434. https://doi.org/10.1038/nnano.2007.180 (2007).
Lee, K.-W. et al. An underlap field-effect transistor for electrical detection of influenza. Appl. Phys. Lett. 96, 033703 (2010).
Okyay, A. K. et al. Using nanogap in label-free impedance based electrical biosensors to overcome electrical double layer effect. Microsyst. Technol.-Micro- Nanosyst.-Inf. Storage Process. Syst. 23(4), 889–897. https://doi.org/10.1007/s00542-015-2764-4 (2017).
Narang, R., Reddy, K. S., Saxena, M., Gupta, R. S. & Gupta, M. A dielectric-modulated tunnel-FET-based biosensor for label-free detection: Analytical modelling study and sensitivity analysis. IEEE Trans. Electron Devices 59(10), 2809–2817. https://doi.org/10.1109/TED.2012.2208115 (2012).
Maji, M. & Saini, G. Modelling of dual material surrounding split gate junctionless transistor as biosensor. Superlattices Microstruct. 135, 106290 (2019).
Bhattacharyya, A., Chanda, M. & De, D. Analysis of partial hybridization and probe positioning on sensitivity of a dielectric modulated junctionless label free biosensor. IEEE Trans. Nanotechnol. 19, 719–727. https://doi.org/10.1109/TNANO.2020.3025544 (2020).
Narang, R., Saxena, M. & Gupta, M. Comparative analysis of dielectric modulated FET and TFET-based biosensor. IEEE Trans. Nanotechnol. 14(3), 427–435 (2015).
Kim, et al. A new sensing metric to reduce data fluctuations in a nanogap-embedded field-effect transistor biosensor. IEEE Trans. Electron Devices 59(10), 2825–2831. https://doi.org/10.1109/TED.2012.2209650 (2012).
Park, H. et al. Label-free and recalibrated multilayer \(\text{ MoS}_{{2}}\) biosensor for point-of-care diagnostics. ACS Appl. Mater. Interfaces 9(50), 43490–43497. https://doi.org/10.1021/acsami.7b14479 (2017).
Pumera, M. & Loo, A. H. Layered transition-metal dichalcogenides (\(\text{ MoS}_{{2}}\) and \(\text{ WS}_{{2}}\)) for sensing and biosensing. TrAC Trends Anal. Chem. 61, 49–53. https://doi.org/10.1016/j.trac.2014.05.009 (2014).
Sarkar, D. et al. MoS2 Field-Effect Transistor for Next-Generation Label-Free Biosensors. ACS Nano. 8(4), 3992–4003 (2014).
Masurkar, N., Varma, S. & Mohana Reddy Arava, L. Supported and suspended 2D material-based FET biosensors. Electrochem 1, 260–277. https://doi.org/10.3390/electrochem1030017 (2020).
Lee, J. et al. Two-dimensional layered \(\text{ MoS}_{{2}}\) biosensors enable highly sensitive detection of biomolecules. Sci. Rep. 4, 7352. https://doi.org/10.1038/srep07352 (2015).
Wen, W. et al. Recent advances in emerging 2-D nanomaterials for biosensing and bioimaging applications. Mater. Today 21(2), 164–177 (2018).
Gan, X., Zhao, H. & Quan, X. Two-dimensional \(\text{ MoS}_{{2}}\): A promising building block for biosensors. Biosens. Bioelectron. 89, 56–71. https://doi.org/10.1016/j.bios.2016.03.042 (2017).
Alam, K. & Lake, R. K. Monolayer \(\text{ MoS}_{{2}}\) transistors beyond the technology road map. IEEE Trans. Electron Devices 59(12), 3250–3254 (2012).
Ilatikhameneh, et al. Tunnel field-effect transistors in 2-D transition metal dichalcogenide materials. IEEE J. Explor. Solid-State Comput. Devices Circuits 1, 12–18. https://doi.org/10.1109/JXCDC.2015.2423096 (2015).
Rahman, E., Shadman, A., Ahmed, I., Khan, S. Z. & Khosru, Q. D. M. Monolayer \(\text{ MoS}_{{2}}\) and \(\text{ WSe}_{{2}}\) double gate field effect transistor as super Nernst pH sensor and nanobiosensor. Sens. Bio-Sens. Res. 11(1), 45–51 (2016).
Jawaid, A. et al. Mechanism for liquid phase exfoliation of \(\text{ MoS}_{{2}}\). Chem. Mater. 28, 337–348. https://doi.org/10.1021/acs.chemmater.5b04224 (2016).
Wang, et al. Functionalized \(\text{ MoS}_{{2}}\) nanosheet-based field-effect biosensor for label-free sensitive detection of cancer marker proteins in solution. Small (Weinheim an der Bergstrasse, Germany) 10(6), 1101–1105. https://doi.org/10.1002/smll.201302081 (2014).
Nam, H. et al. Two different device physics principles for operating \(\text{ MoS}_{{2}}\) transistor biosensors with femtomolar-level detection limits. Appl. Phys. Lett. 107(1), 012105 (2015).
Rahman, E., Shadman, A., Ahmed, I., Khan, S. Z. & Khosru, Q. D. M. A physically based compact I-V model for monolayer TMDC channel MOSFET and DMFET biosensor. Nanotechnology 29(23), 235203 (2018).
Fang, H. et al. High-performance single layered \(\text{ WSe}_{{2}}\) p-FETs with chemically doped contacts. Nano Lett. 12(7), 3788–3792 (2012).
ATLAS User’s Manual. (Silvaco IncA, 2015).
Masurkar, et al. Reliable and highly sensitive biosensor from suspended \(\text{ MoS}_{{2}}\) atomic layer on nanogap electrodes. Biosens. Bioelectronics Retrieved 172, 112724. https://doi.org/10.1016/j.bios.2020.112724 (2021).
Rawat, et al. Two-dimensional \(\text{ MoS}_{{2}}\)-based electrochemical biosensor for highly selective detection of glutathione. IEEE Sens. J. 20(13), 6937–6944. https://doi.org/10.1109/JSEN.2020.2978275 (2020).
Rawat, B., Vinaya, M. M. & Paily, R. Transition metal dichalcogenide-based field-effect transistors for analog/mixed-signal applications. IEEE Trans. Electron Devices 66(5), 2424–2430. https://doi.org/10.1109/TED.2019.2906235 (2019).
Nam, H. et al. Fabrication and comparison of \(\text{ MoS}_{{2}}\) and \(\text{ WSe}_{{2}}\) field-effect transistor biosensors. J. Vacuum Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenomena 33(6), 06FG01. https://doi.org/10.1116/1.4930040 (2015).
Tsividis, Y. & McAndrew, C. Operation and Modelling of the MOS Transistor 2nd edn. (Oxford University, 2011).
Singh, S. & Raja, B. Analytical modelling of split-gate junction-less transistor for a biosensor application. Sens. Bio-Sens. Res. 18, 31–36 (2018).
Kumari, M. et al. Work function optimization for enhancement of sensitivity of dual-material (DM), double-gate (DG), junctionless MOSFET-based biosensor. Appl. Phys. A 127, 130. https://doi.org/10.1007/s00339-020-04256-0 (2021).
Chakraborty, A. & Sarkar, A. Analytical modelling and sensitivity analysis of dielectric-modulated junctionless gate stack surrounding gate MOSFET (JLGSSRG) for application as biosensor. J. Comput. Electron 16, 556–567. https://doi.org/10.1007/s10825-017-0999-2 (2017).
Narang, R., Saxena, M. & Gupta, M. Investigation of dielectric modulated (DM) double gate (DG) junctionless MOSFETs for application as a biosensors. Superlattices Microstruct. 85, 557–572 (2015).
Das, R. et al. Analytical modelling of sensitivity parameters influenced by practically feasible arrangement of bio-molecules in dielectric modulated FET biosensor. Silicon https://doi.org/10.1007/s12633-021-01617-z (2022).
Kumari, M., Singh, N. K., Sahoo, M. & Rahaman, H. 2-D analytical modelling and simulation of dual material, double gate, gate stack engineered, junctionless MOSFET based biosensor with enhanced sensitivity. Silicon https://doi.org/10.1007/s12633-021-01223-z (2021).
Maiti, S., De, A. & Sarkar, S. K. Analytical modelling of symmetric gate underlap quadruple gate multichannel junctionless MOSFET biosensor. Silicon https://doi.org/10.1007/s12633-021-01415-7 (2021).
Singh, K. N. & Dutta, P. K. Analytical modelling of a high-K underlap dielectric- and charge-modulated silicon-on-nothing FET based biosensor. J. Comput. Electron. 19, 1126–1135. https://doi.org/10.1007/s10825-020-01511-8 (2020).
Parija, S. K. et al. Comparison study of DGMOSFET with and without gate stack configuration for biosensor applications. Silicon https://doi.org/10.1007/s12633-021-01118-z (2021).
Cao, W., Kang, J., Liu, W. & Banerjee, K. A compact current-voltage model for 2D semiconductor based field-effect transistors considering interface traps, mobility degradation, and inefficient doping effect. IEEE Trans. Electron Devices 61(12), 4282–4290 (2014).
Jimenez, D., Miranda, E. & Godoy, A. Analytic model for the surface potential and drain current in negative capacitance field-effect transistors. IEEE Trans. Electron Devices 57(10), 2405–2409 (2010).
Bansal, A., Paul, B. C. & Roy, K. Modelling and optimization of fringe capacitance of nanoscale DGMOS devices. IEEE Trans. Electron Devices 52(2), 256–262. https://doi.org/10.1109/TED.2004.842713 (2005).
Mohapatra, N. R., Desai, M. P., Narendra, S. G. & Ramgopal Rao, V. Modelling of parasitic capacitances in deep submicrometer conventional and high-K dielectric MOS transistors. IEEE Trans. Electron Devices 50(4), 959–966. https://doi.org/10.1109/TED.2003.811387 (2003).
Lin, S. C. & Kuo, J. B. Modelling the fringing electric field effect on the threshold voltage of FD SOI nMOS devices with the LDD/sidewall oxide spacer structure. IEEE Trans. Electron Devices 50(12), 2559–2564. https://doi.org/10.1109/TED.2003.816910 (2003).
Francis, P., Terao, A., Flandre, D. & Van de Wiele, F. Modelling of ultrathin double-gate nMOS/SOI transistors. IEEE Trans. Electron Devices 41(5), 715–719 (1994).
Razavi, P. & Orouji, A. A. Nanoscale triple material double gate (TMDG) MOSFET for improving short channel effects. In Proc. ICAE, 11–14 (2008).
Singh, N. K. & Sahoo, M. Analytical modelling of short-channel TMD TFET considering effect of fringing field and 2-D junctions depletion regions. IEEE Trans. Electron Devices 69(2), 843–850 (2022).
Morgan, H., Taylor, D. M. & D’Silva, C. Surface plasmon resonance studies of chemisorbed biotin-streptavidin multilayers. Thin Solid Films 209(1), 122–126 (1992).
Author information
Authors and Affiliations
Contributions
The idea was formulated by M.K. and M.S. M.K. did the simulation and modelling. This paper’s technical analysis was developed by M.K. and N.K.S. All three authors contributed equally to the writing. M.K. and M.S. completed the manuscript’s finalization.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kumari, M., Singh, N.K. & Sahoo, M. A detailed investigation of dielectric-modulated dual-gate TMD FET based label-free biosensor via analytical modelling. Sci Rep 12, 21115 (2022). https://doi.org/10.1038/s41598-022-24677-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-24677-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.