Abstract
Acute-on-chronic liver failure (ACLF) is a critical and refractory disease and a hepatic disorder accompanied by immune dysfunction. Thus, it is essential to explore key immune-related genes of ACLF and investigate its mechanisms. We used two public datasets (GSE142255 and GSE168048) to perform various bioinformatics analyses, including WGCNA, CIBERSORT, and GSEA. We also constructed an ACLF immune-related protein–protein interaction (PPI) network to obtain hub differentially expressed genes (DEGs) and predict corresponding miRNAs. Finally, an ACLF rat model was established to verify the results. A total of 388 DEGs were identified in ACLF, including 162 upregulated and 226 downregulated genes. The enrichment analyses revealed that these DEGs were mainly involved in inflammatory-immune responses and biosynthetic metabolic pathways. Twenty-eight gene modules were obtained using WGCNA and the coral1 and darkseagreen4 modules were highly correlated with M1 macrophage polarization. As a result, 10 hub genes and 2 miRNAs were identified to be significantly altered in ACLF. The bioinformatics analyses of the two datasets presented valuable insights into the pathogenesis and screening of hub genes of ACLF. These results might contribute to a better understanding of the potential molecular mechanisms of ACLF. Finally, further studies are required to validate our current findings.
Similar content being viewed by others
Introduction
Acute-on-chronic liver failure (ACLF) is a major form of liver failure and a chronic liver disease syndrome with acute decompositions that presents a rapid progression and high mortality. The clinical symptoms of ACLF patients include jaundice, ascites, hepatic encephalopathy, or extrahepatic manifestations such as coagulopathy, acute kidney injury, or sepsis1. A meta-analysis that covered 30 cohort studies worldwide showed that the 28- and 90-day mortality rates of ACLF patients were 45 and 58%, respectively2. However, besides liver transplantation, there is no definitive and effective treatment for ACLF. Moreover, the specific treatment for this life-threatening disease has been restricted by the lack of knowledge regarding its molecular mechanisms.
Recently, various bioinformatics approaches have been used to explore the molecular mechanisms of ACLF. Through bioinformatics, massive samples can be processed in a short time to uncover underlying biological mechanisms3. Previous studies have used transcriptome analysis of PMBS, neutrophils, and liver tissues to screen and identify differential genes and obtain characteristic mRNA and miRNA expression profiles for ACLF. They found that ELANE, MPO, and CD177 were upregulated and proposed a miR-6840-3p-JADE2 mRNA-miRNA interaction network that affects the prognosis of ACLF patients4,5. Additionally, a previous genomic study has identified abnormal expression profiles of neutrophils, macrophages, and lymphocytes6. These extensive gene expression reports are of great significance for the study of ACLF. However, specific gene modules with certain biological functions of ACLF have not yet been analyzed, which is equally important for potential ACLF treatment strategies.
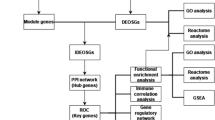
Weighted correlation network analysis (WGCNA) can be used to find co-expressed gene modules and explore the correlation between gene networks and disease phenotypes by assuming that gene networks follow a scale-free distribution and clustering genes with similar expression patterns7. The CIBERSORT algorithm can be used to analyze immune cell composition from complicated tissue gene expression profiles to estimate immune infiltration based on RNA-seq data8. This algorithm applies to gene expression profiles of almost all tissues and covers a wide range of immune cell types, being widely used for estimating the abundance of immune cells in various tissues. In the present study, we used multiple bioinformatics approaches to analyze ACLF-related microarray datasets in the GEO database, identify key immune-related genes, and provide novel strategies for ACLF treatment. We identified the immune cells involved in ACLF, constructed gene co-expression modules associated with these immune cells and hub genes, and predicted reciprocal miRNAs. Finally, the expression of these hub genes and miRNAs was analyzed using an ACLF rat model. Our current findings might provide a valuable reference for the pathogenesis and potential molecular targets of ACLF, and can contribute to the development of therapeutic targets and corresponding drugs for this disease. The workflow of this study is shown in Fig. 1.
Materials and methods
Acquisition and processing of the gene expression matrix
Here, we retrieved clinical samples of ACLF from the GEO database (http://www.ncbi.nlm.nih.gov/geo), a public functional genome database that preserves gene expression and microarray data (Table 1). Twenty-four whole blood samples were selected from GSE142255, comprehending 7 healthy subjects (HS) and 17 ACLF patients. Besides, the GSE168048 dataset was used for validation and described the 28-day survival status of HBV-ACLF patients (8 surviving cases and 8 dead controls). All samples were from Homo sapiens.
Differentially expressed genes (DEGs)
The identification of DEGs was performed using the “limma” R package provided by Sangerbox tools, a free online platform for data analyses (http://www.sangerbox.com/tool). The p-values and adjusted p-values were calculated using t-tests. Genes in each sample that met |log2 (fold change) |> 1 and adjusted p < 0.05 were retained.
Enrichment analyses
Gene Set Enrichment Analysis (GSEA) is a computational method used to determine whether a pre-defined gene set can exhibit consistent significant differences in two biological states9. The c2.cp.kegg.v7.4.symbols.gmt was chosen as a control to assess ACLF-related pathways. Based on gene expression profiles and phenotypic groupings, a minimum gene set of 5 and a maximum gene set of 5000 was selected. One thousand resamplings were performed and a p-value < 0.05 and an FDR < 0.25 were considered statistically significant. Then, to explore which pathways were involved in ACLF pathogenesis, the complete gene expression matrix information of GSE142255 was uploaded to GSEA, and the enrichment results satisfying |NES|≥ 1.0 and NOM p < 0.05 were considered statistically significant. The Metascape10 database and KEGG database (https://www.kegg.jp/kegg/kegg1.html) were used to perform Gene Ontology (GO) and KEGG pathway enrichment analyses by submitting specific gene lists, including upregulated and downregulated DEGs, with min overlap = 3, min enrichment = 1.5, and p < 0.01. Besides, biological processes (BP) networks of DEGs were analyzed and visualized using Cluego in Cytoscape11. The kappa score was set as 0.4. GO terms with at least 9 gene hits and gene percentages > 9% were selected and a p < 0.05 was considered statistically significant.
Immune cell infiltration analysis and WGCNA
CIBERSORT is an algorithm for estimating cellular compositions in tissues based on normalized gene expression data12. In the current study, whole blood mRNA expression profile data from ACLF patients and HS were extracted from GSE142255. CIBERSORT was used to assess the relative proportions of 22 immune cells. The CIBERSORT method was chosen to calculate the immuno-infiltrating cell scores for each sample based on the expression profiles obtained using the IOBR R package. To identify gene co-expression modules, the top 15,000 genes with the most significant differences between ACLF patients and HS were used to construct a co-expression network via the WGCNA package provided by Sangerbox tools13. The proper power was calculated based on the pickSoftThreshold function. Gene profiles were considered as unsigned co-expression networks. The execution parameters were: power = 14, blockSize = 7000, minModuleSize = 20, deepSplit = 2, mergeCutHeight = 0.25, hub cut = 0.9, net threshold = 0. The gene co-expression modules obtained were further used for association analysis with primarily infiltrating immune cells indicated by CIBERSORT. The correlations between modules and immune infiltrative features were analyzed and presented as heat maps. The grey module represents a collection of genes that could not be assigned to any module. Gene Significance (GS) represents the association between gene expression and each feature, while Module Membership (MM) is the correlation of gene expression with each module. The correlation between MM and GS was calculated using Pearson correlation analysis. The identification of gene clusters and the protein–protein interaction (PPI) network analysis were also performed. Intersecting genes were obtained from the most relevant modular DEGs and uploaded to Metascape for PPI network construction, and GO and KEGG pathway enrichment analyses. A p < 0.05 was considered statistically significant.
Screening of hub genes and validation with the GSE168048 dataset
CytoHubba is a Cytoscape plugin used to analyze and acquire key genes in a network and provides various topology analysis algorithms, including Degree, Edge Percolated Component (EPC), Maximum Neighborhood Component (MNC), Density of Maximum Neighborhood Component (DMNC), Maximal Clique Centrality (MCC), Bottleneck (BN), EcCentricity, Closeness, Radiality and Betweenness14. In the present study, each node was ranked using these ten topological analysis algorithms. Using the "UpSet" R package, hub genes were filtered according to the first 60 genes in each algorithm and verified using the GSE168048 dataset.
Construction of a potential miRNA-target regulatory network
We used miRnet 2.0 (https://www.mirnet.ca/) to search for the corresponding miRNAs of hub genes and visualize the miRNA-target regulatory network15. The miRNAs with 2 or more degrees were displayed.
Animals and ACLF model
A total of 18 Wistar rats (180–220 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China. After one week of acclimatization, rats were randomly divided into two groups: normal control (NC, n = 9) and ACLF (ACLF, n = 9). The ACLF rat model was established as previously described. Briefly, rats were intraperitoneally injected with 40% carbon tetrachloride (CCl4) olive oil solution (1.5 ml/kg) twice a week for ten weeks and an acute liver injury was induced by intraperitoneal injection of LPS (100 μg/kg) and D-GalN (400 mg/kg). After 12 h, all rats were sacrificed and the livers were collected and stored at − 80 ℃. Histological observation.
To make tissue sections, rat liver tissues were first embedded in paraformaldehyde, then in paraffin. Processed tissue sections were stained with hematoxylin–eosin (HE) and Masson.
Immunofluorescence staining
Sections were dewaxed with xylene and ethanol, and antigen-repaired with citric acid buffer. Tissues were blocked with 10% goat serum for 1 h and co-incubated with iba1 (1:100, sc-32725, Santa Cruz, USA) and CD68 (1:100, ab283654, Abcam, UK) antibodies at 4 °C. After rinsing with PBST, the sections were incubated with CoraLite488-conjugated Goat Anti-Mouse IgG(H + L) (1:500, SA00013-4, Proteintech, USA) and CoraLite594—conjugated Goat Anti-Rabbit IgG(H + L) (1:500, SA00013-4, Proteintech, USA) for 2 h at 37 °C. After rinsing, tissue sections were covered with an antifade mounting medium with DAPI and observed using a Leica TCS SP8 STED 3X Super-Resolution Confocal Microscope (Leica, Germany). Ten fields of view were randomly selected for each liver section (× 400).
Quantitative real-time PCR (qRT-PCR) analysis
Briefly, the RNA was extracted from livers using Trizol and 50 µL of RNase-free water was added to lyse the RNA. The RNA was reverse transcribed to cDNA by adding primers synthesized by the Beijing Tianyi Huiyuan Biotechnology Co., LTD. Then, the qRT-PCR reaction was performed using the TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) kit mixed with cDNA. The reaction conditions comprehended 40 cycles of 95 °C for 10 s, and 60 °C for 30 s after pre-denaturation for 5 min. The 2−ΔΔCt method was used to analyze the expression of mRNAs and miRNAs. The mRNA expression was normalized to β -actin, while miRNA was normalized to U6. The primer sequences are shown in Table 2.
Ethical approval
All animal experiments were performed according to ARRIVE guidelines and carried out in the Experimental Animal Center of Capital Medical University. All protocols were carried out in accordance with current legislation relating to animals and experiments involving animals and were approved by the Animal Experiments and Experimental Animal Welfare Committee of Capital Medical University (Beijing, China).
Results
Screening of DEGs
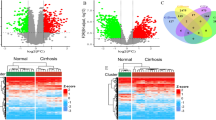
Two microarray datasets (GSE12255 and GSE168048) were used to screen DEGs with the following criteria: adjusted p-value < 0.05 and |log2 FC|> 1. A total of 388 DEGs (226 downregulated and 162 upregulated) were identified after removing unannotated probes and duplicated genes in GSE142255 (Fig. 2A,C, and Supplementary Table S1). Meanwhile, 386 DEGs were upregulated and 223 were downregulated in the GSE168048 dataset (Fig. 2B,D, and Supplementary Table S2).
Visualization of gene expression on GSE142255 and GSE168048. Differentially expressed genes (DEGs) in GSE142255 (A) and GSE168048 (B) were screened with |log2 (fold change) |> 1 and adjusted p < 0.05 and are presented in a Volcano plot. Blue plots represent downregulated DEGs and red plots represent upregulated ones. The expression of DEGs of each sample in GSE142255 (C) and GSE168048 (D) was visualized in clustered heat maps.
Pathway enrichment analysis and functional annotation of GSE142255
All annotated gene information of ACLF and HS in GSE142255 was uploaded to the GSEA software for analysis at a holistic level. Pathways with |NES|≥ 1.0 and NOM p-value < 0.05 were considered as significantly enriched gene sets. The GSEA results revealed significant enrichment in immune-related functions and biosynthesis metabolic pathways (Fig. 3), including primary immunodeficiency, and fructose and mannose metabolism. Using Metascape, 162 upregulated and 226 downregulated DEGs were enriched in GSE142255 according to the KEGG pathway analysis (Supplementary Table S3). Downregulated DEGs were mainly engaged in multiple immune responses and inflammatory signaling pathways, including Th1 and Th2 cell differentiation, T cell receptor signaling pathway, Natural killer cell-mediated cytotoxicity, Chemokine signaling pathway, NF-kappa B signaling pathway, and Apoptosis. Meanwhile, upregulated DEGs affected biosynthetic and substance metabolism pathways, including Starch and sucrose metabolism, MAPK signaling pathway, Fatty acid biosynthesis, Insulin signaling, Cytokine-cytokine receptor interaction, Fructose and mannose metabolism, and Central carbon metabolism in cancer (Fig. 4A,C). These enrichment results validated the GSEA results and were visualized with corresponding DEGs using string diagrams to display the relationship between DEGs and ACLF pathogenesis (Fig. 4B,D).
Moreover, we performed a GO-BP enrichment analysis of DEGs and constructed an interaction network using the ClueGo plug-in in Cytoscape (Fig. 5). The network revealed that the biological processes of ACLF mainly included the activation, differentiation, proliferation, and migration of immune cells.
Identification of gene co-expression modules by WGCNA
The top 15,000 genes in GSE142255 were used to construct a co-expression network with a β power = 14 (Fig. 6A), resulting in 28 gene modules. Genes in the same module have similar expression profiles. Each module contained at least 20 genes with a merge cut height of 0.25. The clustering relationship between modules was represented in a heat map to show the correlation between different modules (Fig. 6B,C).
Gene co-expression modules associated with ACLF were identified by WGCNA. (A) The clustering analysis of ACLF patients and HS in GSE1422552 with a β = 14 as soft threshold (power) and scale-free topology fitting index (R2) = 0.9. (B) Twenty-eight gene co-expression modules were obtained. (C) The relationship between modules was visualized in a heat map.
Correlation analysis of immune infiltration and gene modules
As reported in GSE142255, ACLF patients are characterized by dysregulated blood immune cells, such as neutrophils, and several lymphocyte subsets. Herein, we used the CIBERSORT algorithm to investigate the correlation between immune infiltration and gene co-expression modules. The proportion of each immune cell type in each sample is shown in a stacked bar graph (Fig. 7A). The proportions of M0 and M1 macrophages increased, while the proportion of monocytes was not significantly altered, indicating significant macrophage differentiation and polarization in ACLF (Fig. 7B). Moreover, the coral1 module had significant negative correlations with M0 (r =− 0.63, p < 0.01) and M1 (r =− 0.65, p < 0.01) macrophages. On the other hand, the darkseagreen4 module had a significant positive correlation with M0 (r = 0.7, p < 0.01) and M1 (r = 0.67, p < 0.01) macrophages (Fig. 8).
Correlation analysis between M0 and M1 macrophages, and key gene expression modules. (A) The heat map revealed that variations in M0 and M1 macrophages were associated with coral1 and darkseagreen4 modules, with corresponding correlation and p-value in each cell. (B) The scatter plot revealed that Gene significance (GS) and module membership (MM) are highly correlated in coral1 and darkseagreen4 modules.
Construction and analysis of a PPI network of macrophage-related DEGs
To screen out macrophage-related DEGs, 297 genes were obtained from the intersection of the coral1 and darkseagreen4 modules and DEGs (Fig. 9A). Then, the PPI network was constructed, resulting in 168 nodes, 469 edges, and 7 gene clusters (Fig. 9B,C). The GO annotation and KEGG pathway analyses of the PPI network are shown in Fig. 10 and Supplementary Table S4.
Identification of hub genes
Based on the PPI network, we used ten topological analysis methods provided by CytoHubba to calculate and rank the parameters of each node. Then, ten hub genes were obtained: RSL1D1, RPS5, CCL5, HSPA8, PRKCQ, MMP9, ITGAM, LCK, IL7R, and HP (Fig. 11). Further, we explored the genes on GSE168048. We found that the expression of RPS5, PRKCQ, MMP9, LCK, ITGAM, IL7R, and CCL5 was statistically different between surviving and dead samples (Fig. 12A). The expression remained consistent in both datasets, suggesting that these genes are associated with the 28-day survival status of ACLF patients and might be critical in ACLF.
Prediction of miRNAs related to hub genes
The ten hub genes were submitted to miRNet 2.0 for miRNA prediction and network construction. The miRNAs with degree values ≥ 3 are shown in Fig. 12B. The degree values of Has-mir-1-3p, has-mir-9-5p, has-mir-16-5p, has-mir-182-5p, and has-mir-26a-5p were equal to 4. Hence, they might be crucial miRNAs during ACLF development.
Experimental verification in vivo
We assessed the pathological damage in ACLF model rats by HE, masson staining. The liver lobules of the rats in NC had a regular structure, and there were no necrotic cells, without obvious inflammatory cell infiltration or fibrous hyperplasia. However, the hepatocytes in the ACLF group were disorganized, with large and sub-large pieces of severe necrosis. A large infiltration of inflammatory cells is seen in the visual field, as well as marked hepatic sinusoidal haemorrhage. The hilar region is bridged by proliferating collagen fibres and pseudobullets are formed (Fig. 13A). Furthermore, liver sections were double-stained for iba1 (macrophages marker) and CD68 (M1 marker) by immunofluorescence16,17. A significant increase in iba1 and CD68 positive staining cells was detected in the livers of ACLF rats, suggesting that ACLF macrophages were recruited and induced to M1 polarization (Fig. 13B,C). Consistent with the results predicted by CIBERSORT, a significant macrophage infiltration occurred in ACLF rats. Subsequently, we used qRT-PCR to examine the expression levels of hub genes and miRNAs in ACLF rats compared to controls. The results showed a significant decrease in the mRNA expression of RSL1D1, RPS5, CCL5, HSPA8, PRKCQ, LCK, and HP (p < 0.05, p < 0.01, p < 0.001), while MMP9, ITGAM, and IL7R were significantly increased (p < 0.05, p < 0.01) (Fig. 13D). Then, to validate our predicted miRNAs, the miRNAs homologous to humans were matched using the miRBase database (https://mirbase.org/) and analyzed via qRT-PCR. The expression of mir-16-5p increased (p < 0.05) and mir-26a-5p decreased in ACLF rats (p < 0.05). In contrast, the expression of mir-182 and mir-9a-5p did not differ from controls (Fig. 13E).
Experimental validation by establishing an ACLF rat model. (A) Pathological damage in rat liver observed by HE and Masson staining (Scale bars: 100 μm). (B) Liver sections were co-stained with iba1 (macrophages marker) and CD68 (M1 marker) (Scale bars: 50 μm, × 400). (C) Quantitative analysis of CD68 + /iba1 + macrophages in each field. The expression levels of hub gene (D) as well as miRNA (E) in ACLF rats were measured with qRT-PCR. *p < 0.05, **p < 0.01 and ***p < 0.001.
Discussion
ACLF is a systemic inflammatory disease accompanied by immune dysfunction and disturbances in energy metabolism. ACLF has a high short-term mortality, which increases with the incidence of failing organs. Although many studies regarding ACLF have been performed, its underlying mechanisms remain to be fully explored. Meanwhile, it has been demonstrated that a strong immune response is a key mechanism of ACLF18,19. Therefore, we explored the intrinsic mechanisms of immune cell infiltration in ACLF using an effective informational biology approach.
Herein, we identified macrophage-associated co-expressed gene modules in ACLF for the first time using a combination of WGCNA and CIBERSORT. We identified immune-related key genes and provided new pathways for future studies on effective targets for ACLF treatments. After bioinformatics and qRT-PCR experiments, 10 immune-related hub genes were identified and mir-16-5p and mir-26a-5p were validated. Altogether, these results might provide new strategies for understanding the pathogenesis of ACLF and developing targeted therapeutic molecules.
In the present study, we evaluated potential pathways and biological processes of ACLF using enrichment analyses. GSEA is characterized by the analysis of collections of genes rather than individual genes, which helps to avoid the inability to reproduce individual high-scoring genes due to poor annotation. In the GSE142255 dataset, the GSEA indicated that immune response, inflammatory pathways, and metabolic pathways were mainly involved in ACLF. Then, we found that the downregulated DEGs were mainly engaged in immune response and inflammatory reaction, while upregulated ones regulated biosynthetic and substance metabolism pathways. These results reflected two major biological processes that co-occurred during the progression of ACLF regulated by different genes: imbalance of immune-inflammatory response and energy metabolism. According to the BP analysis, immune cell activation, differentiation, proliferation, and migration were the major biological processes in ACLF, leading to an expanding inflammatory response. Recently, it was reported that excessive activation of the immune response not only causes a systemic inflammatory response, which subsequently mediates immune-related tissue damage, but also leads to high energy demand. Consequently, the immune system competes with peripheral organs for energy, triggering an immune-related energy crisis in the organism and increasing the risk of organ failure18,20,21. Overall, it was suggested that the hyperimmune response and dysfunctional energy metabolism in ACLF are biologically coupled processes, largely influencing ACLF progress.
CIBERSORT is a widely used deconvolution machine algorithm for estimating the composition of immune cells. It shows superior performance in the identification and fine delineation of immune cells when processing highly noisy mixture data8,22. Here, the CIBERSORT results showed that the population of M0 and M1 macrophages was significantly increased in ACLF patients compared to healthy subjects. We also labeled markers on the surface of macrophages by immunofluorescence and validated their increase in M1 macrophages in the liver of ACLF rats. Macrophages can be polarized into M1 or M2 phenotypes. M1 macrophages can release significant influxes of inflammatory factors and induce cytokine storms with pro-inflammatory effects. On the other hand, M2 macrophages secrete tissue repair factors and exhibit anti-inflammatory and reparative properties23,24. Kupffer cells, a type of macrophage that resides in the hepatic sinusoids, mainly perform innate immune and inflammatory responses25. To search for highly related gene modules, WGCNA identifies similar gene clusters and gene modules by hierarchical clustering. WGCNA also supports the analysis of correlations between gene modules and phenotypic traits26,27. To identify gene clusters associated with macrophages, we performed WGCNA and identified gene modules closely related to M1 macrophage polarization, including the coral1 (containing 3631 genes) and darkseagreen4 (containing 307 genes) modules. Based on the WGCNA’s gene modules, we screened immune-related DEGs and constructed a PPI network to find ACLF immune-related hub genes.
Ten hub genes were screened using CytoHubba: RSL1D1, RPS5, CCL5, HSPA8, PRKCQ, MMP9, ITGAM, LCK, IL7R, and HP (Table 3). The differential expression of hub genes was further confirmed by qRT-PCR in ACLF rats. Overall, MMP9, ITGAM, and IL7R were highly expressed during ACLF. Furthermore, ACLF has high 28-day mortality that is closely related to the degree of organ failure in patients. Hence, we used the GSE168048 microarray containing gene expression data of ACLF patients who survived or died at 28 days for further investigation. We verified that the expression of RPS5, PRKCQ, MMP9, LCK, ITGAM, IL7R, and CCL5 differed between surviving and deceased patients, suggesting that these genes might be closely related to ACLF progression and could be used to predict ACLF survival status at 28 days. Notably, downregulated genes were mostly involved in the promotion of immune response, while the upregulated gene, MMP9, was associated with hepatocyte necrosis. These results suggested that the coexistence of immune paralysis and cell necrosis is a potential ACLF mechanism leading to poor prognosis.
Moreover, miRNAs are potential targets in numerous diseases and control various biological processes. As short-chain RNAs with a coding length of only about 22 nucleotides, miRNAs cannot directly be translated into proteins, but rather regulate protein synthesis by disrupting the stability of target mRNAs and inhibiting their translation through complementary pairing28. Studies have explored the relationship between miRNAs and diseases and proposed the use of miRNAs as a biomarker for disease diagnosis and prognosis as well as a small molecule drug target29. Considering the time and cost of experimental studies, we adopted a database approach combined with experimental validation to study miRNAs that were significantly altered in ACLF. The miRNet 2.0 integrates data from 15 prediction databases and provides visual analytics to enable a more comprehensive and convenient evaluation of the interactions between miRNAs, mRNAs, lncRNAs, and transcription factors15. Herein, we used miRNet 2.0 to construct a miRNA-hub genes network to explore potential miRNAs related to ACLF. During the validation, two miRNAs were significantly altered in ACLF rats: mir-16-5p presented increased expression and mir-26a-5p showed decreased expression. M1 macrophages can transfer mir-16-5p to gastric cancer (GC) cells by secreting exosomes and triggering a T-cell immune response to suppress tumor formation by decreasing the expression of PD-L130. It has been demonstrated that mir-26a-5p decreases with ACLF progression and is associated with worsening liver function and increasing liver disease severity31. However, further studies are needed to validate the potential association between miRNA regulatory networks and ACLF.
Predicting potential disease-associated miRNAs is very meaningful and challenging. Thus, researchers have developed several computational methods and models to perform those predictions. These models can be classified into four categories: score functions, complex network algorithms, machine learning, and multiple biological information29. For example, Chen et al.32 proposed an inductive matrix filling model (IMCMDA) for miRNA-disease association prediction. By integrating miRNA and disease similarity information into the matrix-populated objective function, a low-dimensional representation matrix of miRNAs and diseases was obtained, which was finally combined into a miRNA-disease association score matrix. Chen et al.33 improved the HGIMDA model and further provided the MDHGI model. This model first decomposes the miRNA-disease association matrix to remove data noise, then uses the topological information implied to make predictions through heterogeneous graph inference. It combines machine learning with network analysis methods to make effective predictions for new disease-miRNA associations. Further, Chen et al. proposed an Ensemble of Decision Tree-based MiRNA-Disease Association prediction (EDTMDA) model34 based on the construction of multiple decision trees by randomly selecting negative samples, miRNA features, and disease features, and by dimensionality reduction of the features. The mean of the predicted values from these decision trees is used as the miRNA-disease association score. This model incorporates feature dimensionality reduction into integrated learning to remove noise and redundant information in the learning process and reduce the computational complexity of the model with higher prediction accuracy. Moreover, Liu et al.35 proposed a DFELMDA-based deep forest integrated learning approach to infer miRNA-disease correlations. This model trains a random forest by constructing two auto-encoders based on miRNAs and diseases, extracting low-dimensional feature representation, and finally predicting potential miRNA-disease associations through the random forest. This model combines feature and deep forest-integrated learning models to enhance the prediction accuracy. Bioinformatics-based prediction methods are constantly evolving. Nevertheless, different models have almost different predictive performance for the same datasets. Hence, it is not only necessary to collect large-scale experimental data but also consider other algorithms to improve predictive performance for specific diseases.
Besides the methods covered in this study, the multi-field predictive research of bioinformatics offers a unique perspective on the exploration of diagnostic and therapeutic tools for diseases, not only for ACLF. Currently, with the development of genome-wide technologies, there is an increasing need to explore models that detail the exact mechanisms in which genes and proteins interact to form complex living systems. A gene regulatory network (GRN) is a network of interactions between gene molecules. An improved Markov blanket discovery algorithm based on IMBDANET has been proposed and can effectively distinguish between direct and indirect regulatory genes from GN and reduce the false-positive rate in the network inference process36. Additionally, RWRNET is an algorithm of Random Walk with Restart (RWR) modified by restart probability, initial probability vector, and roaming network applied to GRN that continuously maps the global topology of the network and estimates the affinity between nodes in the network through circular iterations until all nodes are traversed37. In contrast, IMBDANET uses a Markov blanket discovery algorithm for network topology analysis and processing, identifying direct and indirect regulatory genes while solving the problem of isolated nodes. On the other hand, RWRNET focuses on global network topology information but it cannot handle isolated nodes. Finally, the integration of different methods can be more beneficial for the prediction of gene regulatory relationships.
Here, we combined WGCNA and CIBERSORT algorithms and employed GSEA, KEGG, and GO enrichment analyses to explore immune-related hub genes and potential biological mechanisms in ACLF. The hub genes and miRNAs involved in ACLF regulation were also further validated. Since there are few studies regarding ACLF mechanisms, adopting bioinformatics analyses provided valid information and guidance for our research. However, our current study also has some limitations. First, we used an animal model rather than samples from humans to validate the ACLF immune-related hub genes, and the results from animal studies should be treated with caution. Furthermore, although these hub genes and miRNAs were altered and might be involved in the development of ACLF, whether these genes can be new therapeutic targets for ACLF still needs to be explored. Therefore, further experiments are required to validate our findings and explore potential ACLF mechanisms.
Conclusion
In summary, we used different bioinformatics approaches to uncover potential ACLF molecular mechanisms. The biological processes and pathways that govern immune activation provided a meaningful insight for studying ACLF pathogenesis. Immune-related hub genes and important miRNAs that might be involved in critical ACLF functions were also identified. Finally, further studies are needed to identify the molecular mechanisms of these key genes, which might also contribute to the understanding of ACLF.
Data availability
The datasets (GSE142255 and GSE168048) analysed during the current study are available in the Gene Expression Omnibus (GEO) repository(http://www.ncbi.nlm.nih.gov/geo/).
Abbreviations
- ACLF:
-
Acute-on-chronic liver failure
- PPI:
-
Protein–protein interaction
- HS:
-
Healthy subjects
- DEGs:
-
Differentially expressed genes
- WGCNA:
-
Weighted correlation network analysis
- GSEA:
-
Gene set enrichment analysis
- GO:
-
Gene ontology
- BP:
-
Biological processes
- MF:
-
Molecular functions
- CC:
-
Cellular components
- GS:
-
Gene significance
- MM:
-
Module membership
- EPC:
-
Edge percolated component
- MNC:
-
Maximum neighborhood component
- DMNC:
-
Density of maximum neighborhood component
- MCC:
-
Maximal clique centrality
- BN:
-
Bottleneck
- OXPHOS:
-
Oxidative phosphorylation
References
Casulleras, M., Zhang, I. W., López-Vicario, C. & Clària, J. Leukocytes, systemic inflammation and immunopathology in acute-on-chronic liver failure. Cells 9(12), 2632. https://doi.org/10.3390/cells9122632 (2020).
Mezzano, G. et al. Global burden of disease: acute-on-chronic liver failure, a systematic review and meta-analysis. Gut https://doi.org/10.1136/gutjnl-2020-322161 (2021).
Pezoulas, V. C. et al. Machine learning approaches on high throughput NGS data to unveil mechanisms of function in biology and disease. Cancer Genom. Proteom. 18, 605–626 (2021).
Ma, S. et al. Identification of a potential miRNA-mRNA regulatory network associated with the prognosis of HBV-ACLF. Front. Mol. Biosci. 8, 657631. https://doi.org/10.3389/fmolb.2021.657631 (2021).
Saha, R. et al. Inflammatory signature in acute-on-chronic liver failure includes increased expression of granulocyte genes ELANE, MPO and CD177. Sci. Rep. 11, 18849. https://doi.org/10.1038/s41598-021-98086-6 (2021).
Weiss, E. et al. Characterization of blood immune cells in patients with decompensated cirrhosis including ACLF. Front. Immunol. 11, 619039. https://doi.org/10.3389/fimmu.2020.619039 (2020).
Kakati, T., Bhattacharyya, D. K., Barah, P. & Kalita, J. K. Comparison of methods for differential co-expression analysis for disease biomarker prediction. Comput. Biol. Med. 113, 103380. https://doi.org/10.1016/j.compbiomed.2019.103380 (2019).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. https://doi.org/10.1038/s41467-019-09234-6 (2019).
Bindea, G. et al. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. https://doi.org/10.1093/bioinformatics/btp101 (2009).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782. https://doi.org/10.1038/s41587-019-0114-2 (2019).
Zhao, W. et al. Weighted gene coexpression network analysis: State of the art. J. Biopharm. Stat. 20, 281–300. https://doi.org/10.1080/10543400903572753 (2010).
Zhou, S., Lu, H. & Xiong, M. Identifying immune cell infiltration and effective diagnostic biomarkers in rheumatoid arthritis by bioinformatics analysis. Front. Immunol. 12, 726747. https://doi.org/10.3389/fimmu.2021.726747 (2021).
Chang, L., Zhou, G., Soufan, O. & Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucl. Acids Res. 48, W244–W251. https://doi.org/10.1093/nar/gkaa467 (2020).
Wijesundera, K. K. et al. M1- and M2-macrophage polarization in rat liver cirrhosis induced by thioacetamide (TAA), focusing on Iba1 and galectin-3. Exp. Mol. Pathol. 96, 382–392. https://doi.org/10.1016/j.yexmp.2014.04.003 (2014).
Tsuji, Y. et al. Acetaminophen-induced rat hepatotoxicity based on M1/M2-macrophage polarization, in possible relation to damage-associated molecular patterns and autophagy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21238998 (2020).
Moreau, R. et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J. Hepatol. 72, 688–701. https://doi.org/10.1016/j.jhep.2019.11.009 (2020).
Kumar, K. & Kulkarni, A. Acute-on-chronic liver failure. N. Engl. J. Med. 383, 892–893. https://doi.org/10.1056/NEJMc2023198 (2020).
Engelmann, C., Claria, J., Szabo, G., Bosch, J. & Bernardi, M. Pathophysiology of decompensated cirrhosis: Portal hypertension, circulatory dysfunction, inflammation, metabolism and mitochondrial dysfunction. J. Hepatol. 75(Suppl 1), S49–S66. https://doi.org/10.1016/j.jhep.2021.01.002 (2021).
Arroyo, V. et al. The systemic inflammation hypothesis: Towards a new paradigm of acute decompensation and multiorgan failure in cirrhosis. J. Hepatol. 74, 670–685. https://doi.org/10.1016/j.jhep.2020.11.048 (2021).
Le, T., Aronow, R. A., Kirshtein, A. & Shahriyari, L. A review of digital cytometry methods: estimating the relative abundance of cell types in a bulk of cells. Brief. Bioinform. https://doi.org/10.1093/bib/bbaa219 (2021).
Sun, Y. Y. et al. Macrophage phenotype in liver injury and repair. Scand. J. Immunol. 85, 166–174. https://doi.org/10.1111/sji.12468 (2017).
van der Heide, D., Weiskirchen, R. & Bansal, R. Therapeutic targeting of hepatic macrophages for the treatment of liver diseases. Front. Immunol. 10, 2852. https://doi.org/10.3389/fimmu.2019.02852 (2019).
Triantafyllou, E., Woollard, K. J., McPhail, M. J. W., Antoniades, C. G. & Possamai, L. A. The role of monocytes and macrophages in acute and acute-on-chronic liver failure. Front. Immunol. 9, 2948. https://doi.org/10.3389/fimmu.2018.02948 (2018).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9, 559. https://doi.org/10.1186/1471-2105-9-559 (2008).
Langfelder, P. & Horvath, S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J. Stat. Softw. 46 (2012).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. https://doi.org/10.1038/nrg1379 (2004).
Chen, X., Xie, D., Zhao, Q. & You, Z. H. MicroRNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 20, 515–539. https://doi.org/10.1093/bib/bbx130 (2019).
Li, Z. et al. Exosomal miRNA-16-5p derived from M1 macrophages enhances T cell-dependent immune response by regulating PD-L1 in gastric cancer. Front. Cell Dev. Biol. 8, 572689. https://doi.org/10.3389/fcell.2020.572689 (2020).
Blaya, D. et al. Profiling circulating microRNAs in patients with cirrhosis and acute-on-chronic liver failure. JHEP Rep. 3, 100233. https://doi.org/10.1016/j.jhepr.2021.100233 (2021).
Chen, X., Wang, L., Qu, J., Guan, N. N. & Li, J. Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics 34, 4256–4265. https://doi.org/10.1093/bioinformatics/bty503 (2018).
Chen, X., Yin, J., Qu, J. & Huang, L. MDHGI: Matrix decomposition and heterogeneous graph inference for miRNA-disease association prediction. PLoS Comput. Biol. 14, e1006418. https://doi.org/10.1371/journal.pcbi.1006418 (2018).
Chen, X., Zhu, C. C. & Yin, J. Ensemble of decision tree reveals potential miRNA-disease associations. PLoS Comput. Biol. 15, e1007209. https://doi.org/10.1371/journal.pcbi.1007209 (2019).
Liu, W. et al. Identification of miRNA-disease associations via deep forest ensemble learning based on autoencoder. Brief. Bioinform. https://doi.org/10.1093/bib/bbac104 (2022).
Liu, W. et al. Inferring gene regulatory networks using the improved markov blanket discovery algorithm. Interdiscip. Sci. 14, 168–181. https://doi.org/10.1007/s12539-021-00478-9 (2022).
Liu, W. et al. RWRNET: A gene regulatory network inference algorithm using random walk with restart. Front. Genet. 11, 591461. https://doi.org/10.3389/fgene.2020.591461 (2020).
Ma, L. et al. Ribosomal L1 domain and lysine-rich region are essential for CSIG/ RSL1D1 to regulate proliferation and senescence. Biochem. Biophys. Res. Commun. 469, 593–598. https://doi.org/10.1016/j.bbrc.2015.12.004 (2016).
Ma, L. et al. CSIG inhibits PTEN translation in replicative senescence. Mol. Cell Biol. 28, 6290–6301. https://doi.org/10.1128/MCB.00142-08 (2008).
Bhat, P. et al. The beta hairpin structure within ribosomal protein S5 mediates interplay between domains II and IV and regulates HCV IRES function. Nucleic Acids Res. 43, 2888–2901. https://doi.org/10.1093/nar/gkv110 (2015).
Li, M. et al. CCL5 deficiency promotes liver repair by improving inflammation resolution and liver regeneration through M2 macrophage polarization. Cell Mol. Immunol. 17, 753–764. https://doi.org/10.1038/s41423-019-0279-0 (2020).
Triantafilou, K., Triantafilou, M. & Dedrick, R. L. A CD14-independent LPS receptor cluster. Nat. Immunol. 2, 338–345. https://doi.org/10.1038/86342 (2001).
Chand, S., Mehta, N., Bahia, M. S., Dixit, A. & Silakari, O. Protein kinase C-theta inhibitors: A novel therapy for inflammatory disorders. Curr. Pharm. Des. 18, 4725–4746. https://doi.org/10.2174/138161212802651625 (2012).
Pfeifhofer-Obermair, C. et al. Role of PKCtheta in macrophage-mediated immune response to Salmonella typhimurium infection in mice. Cell Commun. Signal. 14, 14. https://doi.org/10.1186/s12964-016-0137-y (2016).
Teng, L. et al. Matrix metalloproteinase-9 as new biomarkers of severity in multiple organ dysfunction syndrome caused by trauma and infection. Mol. Cell Biochem. 360, 271–277. https://doi.org/10.1007/s11010-011-1066-0 (2012).
Ohashi, N. et al. Matrix metalloproteinase-9 contributes to parenchymal hemorrhage and necrosis in the remnant liver after extended hepatectomy in mice. World J. Gastroenterol. 18, 2320–2333. https://doi.org/10.3748/wjg.v18.i19.2320 (2012).
DiScipio, R. G., Daffern, P. J., Schraufstatter, I. U. & Sriramarao, P. Human polymorphonuclear leukocytes adhere to complement factor H through an interaction that involves alphaMbeta2 (CD11b/CD18). J. Immunol. 160, 4057–4066 (1998).
Losse, J., Zipfel, P. F. & Jozsi, M. Factor H and factor H-related protein 1 bind to human neutrophils via complement receptor 3, mediate attachment to Candida albicans, and enhance neutrophil antimicrobial activity. J. Immunol. 184, 912–921. https://doi.org/10.4049/jimmunol.0901702 (2010).
Herb, M., Gluschko, A. & Schramm, M. LC3-associated phagocytosis initiated by integrin ITGAM-ITGB2/Mac-1 enhances immunity to Listeria monocytogenes. Autophagy 14, 1462–1464. https://doi.org/10.1080/15548627.2018.1475816 (2018).
Farag, A. K. et al. Novel LCK/FMS inhibitors based on phenoxypyrimidine scaffold as potential treatment for inflammatory disorders. Eur. J. Med. Chem. 141, 657–675. https://doi.org/10.1016/j.ejmech.2017.10.003 (2017).
Luo, T. et al. Lck inhibits heat shock protein 65-mediated reverse cholesterol transport in T cells. J. Immunol. 197, 3861–3870. https://doi.org/10.4049/jimmunol.1502710 (2016).
Barata, J. T., Durum, S. K. & Seddon, B. Flip the coin: IL-7 and IL-7R in health and disease. Nat. Immunol. 20, 1584–1593. https://doi.org/10.1038/s41590-019-0479-x (2019).
Andersen, C. B. F. et al. Haptoglobin. Antioxid. Redox Signal. 26, 814–831. https://doi.org/10.1089/ars.2016.6793 (2017).
Acknowledgements
Authors thank CureEdit for the language polishing services.
Funding
This study was supported by National Key R&D Program of China (Grant No. 2018YFC1704102), Beijing TCM Science & Technology Development Foundation (JJ-2020–51) and National Natural Science Foundation of China (82074237).
Author information
Authors and Affiliations
Contributions
Y.G., Y.D. and Q.Z. contributed to the study design and supervised the study. J.L., X.W., W.H. and H.W. carried out the data preparation. J.L. conducted the study design, performed the data analysis, and provided the visualization. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liang, J., Wei, X., Hou, W. et al. Bioinformatics analyses of potential ACLF biological mechanisms and identification of immune-related hub genes and vital miRNAs. Sci Rep 12, 14052 (2022). https://doi.org/10.1038/s41598-022-18396-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-18396-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.