Abstract
Human cerebellum consists of high density and complexity of neurons. Thus, it is challenging to differentiate cerebellar-like organoids with similar cellular markers and function to the human brain. Our previous study showed that the combination of retinoic acid (RA), Wingless/integrated (Wnt) activator, and Sonic Hedgehog (SHH) activator promotes cerebellar differentiation from human induced pluripotent stem cells (hiPSCs). This study examined phenotypic, metabolic, and biogenesis in early cerebellar development. Cerebellum spheroids were differentiated from human iPSK3 cells. During day 7–14, RA and Wnt activator CHIR99021 were used and SHH activator purmorphamine (PMR) was added later to promote ventralization. Gene expression for early cerebellar layer markers, metabolism, and extracellular vesicle (EV) biogenesis were characterized. Zinc-induced neurotoxicity was investigated as a proof-of-concept of neurotoxicity study. Flow cytometry results showed that there was no significant difference in NEPH3, PTF1A, OLIG2, and MATH1 protein expression between RCP (RA-CHIR-PMR) versus the control condition. However, the expression of cerebellar genes for the molecular layer (BHLE22), the granule cell layer (GABRB2, PAX6, TMEM266, KCNIP4), the Bergmann glial cells (QK1, DAO), and the Purkinje cell layer (ARHGEF33, KIT, MX1, MYH10, PPP1R17, SCGN) was significantly higher in the RCP condition than the control. The shift in metabolic pathways toward glycolysis was observed for RCP condition. The EV biogenesis marker expression was retained. Mild zinc-induced neurotoxicity may exist when zinc exposure exceeds 1.0 µM. RCP treatment can promote specific cerebellar-like differentiation from hiPSCs indicated by gene expression of early cerebellar markers and regionally enriched genes. The higher cerebellar marker expression is accompanied by the elevated glycolysis with the retained EV biogenesis. This study should advance the understanding of biomarkers during early cerebellar development for cerebellum organoid engineering and neurotoxicity study.
Similar content being viewed by others
Introduction
Cerebellum locates in the hindbrain region and consists of highly dense and complex neuron networks in the human brain1. It is challenging to derive human brain cerebellar-like spheroids/organoids with similar biomarkers and function from human pluripotent stem cells (hPSCs), including both human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells2,3. Regulating anterior–posterior (A-P) and dorsal–ventral (D-V) axes is the key for successful cerebellum differentiation4. Fibroblast growth factors (FGFs), Wnts, and retinoic acid (RA) are small molecules that regulate the A-P patterning, whereas those affecting the D–V patterning include Wnts, bone morphogenetic proteins (BMPs), and Sonic Hedgehog (SHH)5. CHIR99021 (CHIR) is a morphogen that activates Wnt pathway, and it modulates neuroepithelia differentiation in a dose-dependent manner. High concentrations of CHIR favors the hindbrain development6 as well as the caudalization factor RA. As a SHH activator, purmorphamine (PMR) promotes ventralization of the brain spheroids7. Cerebellum development starts from the formation of isthmic organizer stimulated by FGF28, which prevents the forebrain differentiation pathway. FGF19 facilitates the self-organization of rostral hindbrain structures with a D-V polarity5, and the ventral identity is enhanced by PMR later. Stroma cell-derived factor (SDF) 1α treatment promotes generation of rhombic lip-like structure as well as laminated cerebellar plate structures9. Sequential utilization of the above small molecules successfully generates cerebellar spheroids with molecular layer, Purkinje cell layer, and granule cell layer as shown in our previous work2, but only a few markers representing the three layers were studied. A close scrutinization of cerebellar biomarkers, metabolic, and biogenesis properties of hPSC-derived cerebellar spheroids would advance our knowledge about early cerebellum development.
Metabolic pathways can shift during hPSC neural differentiation10 and adult stem cell aging and thus became an important indicator to be monitored during stem cell differentiation11,12. The progression from undifferentiated hPSCs, immature progenitor cells, to mature differentiated cells may shift the metabolism from glycolysis to oxidative phosphorylation (OXPHOS) pathway13. In glucose metabolism of hPSCs, the entry of pyruvate into the tricarboxylic acid cycle is inhibited by reduced activity of pyruvate dehydrogenase (PDH) by pyruvate dehydrogenase kinase (PDK). Pyruvate is reduced to replenish nicotinamide adenine dinucleotide to maintain glycolytic activity and pyruvate entering the mitochondria is converted to acetyl-CoA utilized for citrate production13. During hPSC early differentiation, a metabolic switch controlling histone acetylation produces acetyl-CoA through glycolysis which rapidly induces hPSC differentiation14. In human neural stem cell differentiation into motor neurons, a switch from glycolysis to OXPHOS was observed, and mitochondrial biogenesis but not mitochondrial mass was increased15. However, the phenotypic correlation of cerebellar differentiation of hPSCs with the metabolic status has not been well investigated.
Extracellular vesicles (EVs) especially exosomes play important roles in cell–cell communications during stem cell differentiation16,17. Exosomes are nanosized (< 150 nm) membrane vesicles that contain various proteins and microRNAs that regulate cell singling networks. EV biogenesis could be affected by cytoskeleton alterations, hypoxia condition, and mechanical stress, and may exhibit different activities for different cell types18,19,20. Our previous study evaluated the EV biogenesis for forebrain cortical spheroids derived from hiPSCs21,22,23. The EV biogenesis of hindbrain cerebellar spheroids derived from hiPSCs and the effect of the cellular phenotype on EV biogenesis marker expression remains unknown.
In particular, our previous study reported the modulation of Retinoid (using retinoic acid-R), Wnt (using CHIR99021-C), and Sonic Hedgehog (using purmorphamine-P) pathways to promote cerebellar spheroid differentiation from hiPSCs2. Going one step further, this study characterized the derived cerebellar spheroids using RCP treatment on three aspects: (1) early cerebellar biomarkers based on Human Protein Atlas; (2) metabolic pathway shift during cerebellar differentiation; and (3) EV biogenesis marker expression to elucidate paracrine signaling during cell–cell interactions. The goal of this study is to identify in-process control markers that can predict the cerebellar differentiation outcome in a quantitative way. Hindbrain cerebellum spheroids/organoids derived from this study can be used to generate assembloids with isogenic forebrain cerebral organoids24,25,26,27, to study the heterogeneity of microglia28 and blood brain barrier29 in different regions of human brain30, and to investigate the neurotoxicity such as those induced by zinc as shown in this study31,32,33,34.
Results
The protein expression of the cerebellar markers
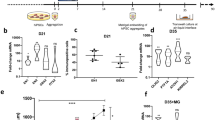
The schematic illustration and morphology of cerebellar differentiation from hiPSCs are represented in Fig. 1. For the control condition, SB431542, FGF2, FGF19 and SDF1α were used as reported previously35. For the RCP tested condition, RA and CHIR were added on week two of culture, and PMR was added on week five. The RCP treatment can induce further caudalization of the spheroids, and therefore, it might promote hindbrain gene expression. NEPH3 and PTF1A, OLIG2, and MATH1 are chosen as the markers for the molecular layer, the Purkinje cell layer, and the granule cell layer of the cerebellar spheroids, respectively. β-tubulin III was used as the general neuronal marker. The immunostaining results, the spheroid maturation by human mesenchymal stem cells, as well as the electrophysiology results have been shown in our previous study2. This study focuses on the quantitative analysis such as flow cytometry and reverse transcription-quantitative polymerase chain reactions (RT-PCR) for cerebellar marker expression and the correlation with the metabolic pathway and EV biogenesis. Flow cytometry results show that there is no significant difference in protein expression of these markers between RCP versus the control or RCP versus FGF8 groups (Fig. 2A,B; Supplementary Table S1)36. Western blot analysis of NEPH3 and OLIG2 protein did not show significant difference between the RCP condition versus the control either (Supplementary Fig. S1). Taken together, RCP treatment does not affect cerebellar marker expression at protein levels for the common markers NEPH3, PTF1A, OLIG2, and MATH1, probably due to large experimental variations in cerebellar spheroid differentiation and flow cytometry sample preparation. In the following study, the new cerebellar marker expression, selected based on Human Protein Atlas on the cerebellum, was performed focusing on the comparison of RCP versus the control condition at a molecular level.
Protocols and morphology of cerebellar differentiation from hiPSCs. (A) Schematic illustration of the cerebellar differentiation from hiPSCs under different conditions. The control condition uses the factors written in the black font. The tested conditions are the combination of RA/CHIR/PMR (RCP) and FGF8 only. (B) Morphology of cerebellar spheroids during differentiation for 35 days. Scale bar: 200 µm.
Flow cytometry analysis for the expression of cerebellar markers in the derived cerebellar spheroids. The control condition for the side-by side-comparison with RCP condition is refereed as Ctrl-RCP. The control condition for the side-by side-comparison with FGF8 condition is refereed as Ctrl-FGF8. (A) Day 35 RCP condition versus the side-by-side control (Ctrl-RCP) or FGF8 condition versus the side-by-side control (Ctrl-FGF8); (B) RCP versus FGF8 condition. The average value and the standard deviation are based on the results from three independent differentiations (each differentiation has the corresponding control condition).
The expression of new human cerebellar specific markers
According to the Human Protein Atlas37,38, 79% of human transcriptome are expressed in the cerebellum, and 13,188 of all genes detected in the brain are detected in cerebellum. Regionally enriched genes in the cerebellum are those that are at least four-fold higher mRNA levels in cerebellum compared to all other regions. There are 214 regionally enriched genes. In addition, regional specificity score (RS-score) corresponds to the score calculated as the fold change to the second highest region. The twelve genes with highest RS-score were selected for RT-PCR analysis in this study to compare the RCP and control conditions, including SPINK6, BARHL2, CCDC155, FGF3, GABRA6, and FAT2 for RS > 30, PRR35, CDH15, CRTAM, ZIC5, CBLN3, and SLC22A31 for 30 > RS > 20 (Supplementary Table S2 and S3). Our results show that the RCP condition can induce a significantly higher gene expression for most markers (10 out of 12) with the exception of FGF3 (RS-score = 38) and ZIC5 (RS-score = 24) compared to the control condition (Fig. 3A,B). The increase is ~ 2 to 7 fold for makers of RS score > 30 and 2–3 fold for markers of 30 > RS > 20. Therefore, RCP treatment can promote cerebellar differentiation from the hiPSCs at the molecular level.
The expression of the top twelve genes with highest RS-score in the human cerebellum and cerebellar layer-specific markers. RT-PCR analysis was performed for day 35 cerebellar organoids derived from hiPSCs. (A) The relative expression of genes (mRNA) with RS-score greater than 30. (B) The relative expression of genes (mRNA) with RS-score between 30 and 20. The expression of cerebellar layer-specific markers. (C) The relative gene (mRNA) expression for the molecular layer, the granule cell layer, and the Bergmann glial cells; (D) the relative gene (mRNA) expression for the Purkinje cell layer. *p < 0.05 compared to the control condition.
Moreover, 14 selected markers for specific cerebellar layer were examined (Supplementary Table S4 and S5). BHLE22 was chosen as the marker for molecular layer. For granule cell layer, GABRB2, PAX6, TMEM266, and KCNIP4 were examined. QK1 and DAO were used as the marker for Bergmannglia cells. For Purkinje cell layer, ARHGEF33, HOMER3, KIT, MX1, MYH10, PPP1R17, and SCGN were determined. The expression of cerebellar specific genes for the molecular layer, the Purkinje cell layer (except HOMER3), and the granule cell layer was significantly higher (~ 2 to 15 fold) in the RCP condition compared with the control (Fig. 3C,D). Interestingly, the expression for the Bergmann glial cells, an important cell type in the cerebellum, is also higher (~ 2 to 6 fold). Therefore, RCP treatment can promote specific cerebellar layer differentiation in spheroids derived from hiPSCs at the molecular level. Selected markers were evaluated for the expression at protein levels by flow cytometry and immunocytochemistry, including FAT2, GABRA6, HOMER3, and KCNIP4 (Fig. 4). No significant difference in protein expression of these markers was observed between RCP versus the control condition.
The expression of cerebellar layer-specific markers at protein levels. Four markers were selected for measuring the expression at the protein levels. (A) Flow cytometry histograms, red line: negative control; blue line: marker of interest. (B) Percentages of positive marker expression quantified by flow cytometry; (C) Immuno-fluorescent images. Scale bar: 100 µm. Blue: Hoechst 333342 for counterstaining with cell nuclei.
Metabolic pathways are altered by RCP treatment
To understand the cerebellar differentiation process, the status of metabolic pathways during differentiation was examined by RT-PCR for day 14 and day 35 cells (Fig. 5A,B; Supplementary Table S6). PDK1 is pyruvate dehydrogenase kinases, which shunts pyruvate away from the mitochondria. HK2 is hexokinase 2, which phosphorylates glucose to glucose 6-phosphate. PKM2 is pyruvate kinase isoform 2, which catalyzes phosphoenolpyruvate (PEP) to pyruvate. LDHA is lactate dehydrogenase A, which catalyzes inter-conversion of pyruvate and lactate. TKTL1 is transketolase-like protein 1, which enables O2-independent glucose degradation. 6PGD is 6-phosphogluconase dehydrogenase responsible for NADPH production. G6PD is glucose-6-phosphate dehydrogenase responsible for NADPH production. TALDO1 is transaldolase 1, which provides ribose-5-phosphate for nucleic acid synthesis and NADPH for lipid biosynthesis.
Characterizations of metabolic pathways during cerebellar differentiation of hiPSCs. RT-PCR analysis of genes (mRNA) related to glycolytic pathway and the pentose phosphate (PPP) pathway was performed for (A) Day 14 cells; (B) Day 35 cells. (i) Glycolytic pathway; (ii) PPP pathway. Spent media were collected for metabolite analysis (glucose, lactate, glutamine, and ammonia). (C) (i) Glucose consumption; (ii) Lactate generation; (iii) Lactate to glucose ratio (mol/mol). *p < 0.05 compared to the control condition.
For glycolytic pathway: at day 35, glycolytic genes (HK2, PKM2, and LDHA) in the RCP group have higher expression levels (~ 1.5 fold) than those in control group, which is different from what was observed for day 14 (~ 1.5 fold lower for PDK1, HK2, and PKM2). For pentose phosphate (PPP) pathway: at both day 14 and day 35, PPP genes (TKTL1, 6PGD, G6PD, and TALDO1) in the RCP group have lower expression levels than those in the control group (2–5 fold higher at day 14 for all the four markers; 1.2–2 fold for TKTL1 and 6PGD only). Our previous study showed that gene expression in metabolic pathways usually correlates with the metabolic enzyme function11. It was postulated that the RCP group has higher oxidative activity to produce more energy for activating genes involved in differentiation at day 14. At day 35, more cerebellar cells in the RCP group increase their use of glycolytic pathway to counteract oxidative stress.
Metabolite analysis (including glucose, lactate, glutamine, and ammonia as well as Na+, K+, Ca2+) was performed on the spent media of cerebellar differentiation (Fig. 5C; Supplementary Fig. S2). Glucose consumption and lactate generation was similar for the control an RCP conditions before day 20. After day 20, glucose consumption and lactate generation were significantly higher for the RCP condition than the control. The lactate to glucose ratio (mol/mol) was around 1.8 for the RCP condition, indicating more anaerobic metabolism (with ratio of 2.0). However, for the control condition, the ratio was lower around 1.0–1.6, especially during day 20–35, indicating more aerobic metabolism. The difference in glutamine metabolism was consistent with glucose metabolism but was shown after day 25. Na+, K+, Ca2+ levels were comparable for the two conditions. Together, these observations were consistent with the results from gene expression.
RC and RCP treatment retains EV biogenesis markers
The EV biogenesis markers are classified as the endosomal sorting complex required for transport (ESCRT) dependent (ALIX, HRS, STAM1, TSG101) and ESCRT independent (CD63, MITF, Rab27b, SMPD2)39. In this study, RT-PCR analysis was performed for day 14 and day 35 cells during cerebellar spheroid differentiation (Fig. 6; Supplementary Table S7). ALIX is involved in exosome generation and cargo loading. HRS, STAM1, TSG101 are related to endosomal sorting complex required for transport. CD63 is related to exosome generation and particle packaging into exosomes. MITF is involved in exosome generation and can increase the expression of late endosomal proteins such as Rab27b. Rab27b facilitates the docking to the plasma membrane. SMPD2 facilitates the budding from the plasma membrane.
Extracellular vesicle (EV) biogenesis marker expression during cerebellar differentiation. The expression levels of EV biogenesis markers were determined by RT-PCR analysis. (A) Day 14 cells; (B) Day 35 cells. (i) ESCRT dependent: ALIX, HRS, STAM1, TSG101; (ii) ESCRT independent: CD63. MITF, Rab27b, SMPD2. *p < 0.05 compared to the control condition. ESCRT: the endosomal sorting complex required for transport. (C) (i) Extracellular vesicle (EV) size distribution determined by nanoparticle tracking analysis; EVs were isolated around day 14 (week 2–3) and around day 35 (week 5–6); (ii) Average mean and mode EV size; and (iii) protein content per 109 EVs. *p < 0.05 compared to other conditions.
At day 14, ALIX, TSG101, and CD63 show the decreased expression (~ 2-fold) for the RCP group compared to the control, while the other five genes show no significant difference between the control group and the RCP group. At day 35, SMPD2 and HRS exhibited the decreased expression (~ 2-fold) for the RCP condition compared to the control, while the other six genes showed the similar expression. The genes having lower expression are not consistent between day 14 and day 35 cells, and they are not exclusively related to ESCRT-dependent or independent category. Therefore, the extent of maturity of cerebellar cells in the derived spheroids does not seem to affect EV biogenesis. The EV biogenesis markers can be evaluated semi-quantitatively using Western blot as shown in our previous study and higher expression of EV biogenesis markers usually correlates with higher EV production or enriched exosome population21,23.
The EV isolation and nanoparticle tracking analysis (NTA) were then performed at the time points around day 14 (media collection at week 2–3) and day 35 (media collection at week 5–6) for the control and RCP conditions (Fig. 6C). NTA results showed that the mode size (120–157 nm) of the isolated EVs were comparable for the two conditions at the two time points. The mode size is usually a more accurate representation of the EV size as vesicle aggregation can affect the mean size. The particle concentrations (2.11–2.80 × 109/mL for the control, and 2.49–3.85 × 109/mL for the RCP condition) (Supplementary Table S8) and the protein content in the EVs were comparable for the control and RCP conditions at week 5–6, although the protein content at week 2–3 for the RCP condition was slightly lower. These observations in general were consistent with the results of gene expression.
Zinc-induced neurotoxicity
As a proof-of-concept study for the use of the derived cerebellar spheroids in neurotoxicity assessment, the derived day 35 cerebellar organoids were exposed to 0, 0.01, 0.1, 1.0, 10 and 100 µM Zinc supplemented culture media (Fig. 7). LIVE/DEAD assay was performed to examine the cell viability. From the images, majority of the cells exposed to the different zinc concentrations are live cells (Fig. 7A). Quantification of percentage of live cells over total cells showed the slight decrease in cell viability for 1.0 and 10 µM exposure compared to the control (Fig. 7B(i)). MTT assay was used to examine the cell metabolic activity. The MTT activity was found to reach the peak for 1.0 µM exposure and the decrease was observed for 10 and 100 µM exposure (Fig. 7B(ii)). LDH assay was performed for determining cytotoxicity. No significant differences were observed for different concentrations of zinc exposure (Fig. 7B(iii)). Taken together, mild zinc-induced neurotoxicity may exist when zinc exposure exceeds 1.0 µM.
Zinc-induced neurotoxicity. (A) Images of LIVE/DEAD assay for the derived cerebellar cells exposed to Zinc at different concentrations. Scale bar: 25 µm. (B) (i) Relative percentage of live cells for the cells exposed to Zinc compared to the control. (ii) Relative MTT activity of cerebellar cells exposed to Zinc at different concentrations compared to the control. (iii) Relative LDH activity of cells exposed to Zinc compared to the control. n = 3. *p < 0.05 compared to the 0.01 µM condition. #p < 0.05 compared to the peak condition.
Discussion
In this study, the gene expression of different cerebellar markers along with metabolic pathway and EV biogenesis markers was determined using RT-PCR to characterize the cerebellar differentiation from hiPSCs by modulating RA, Wnt, and SHH pathways. Compared to forebrain cortical spheroids/organoids derived from hiPSCs, much less publications reported the derivation of hindbrain cerebellum spheroids/organoids26. Cerebellar differentiation from hPSCs has been reported recently in a PBS Vertical Wheel bioreactor based on the control condition (the Muguruma protocol 2015)35. The bioreactor culture accelerates the cerebellar differentiation, angiogenesis, and extracellular matrix based on the transcriptomic analysis2,40. The cerebellar maturation in the absence of co-culture was also reported9. For hindbrain patterning, retinoic acid and Wnt promotes the caudalization of the brain organoids and the activation of sonic hedgehog signaling induces ventralization of the brain tissues41,42,43. Our previous study showed the promoted cerebellar differentiation from hiPSCs by activating retinoid (using retinoic acid-R), Wnt (using CHIR99021-C), and Sonic Hedgehog (using purmorphamine-P) pathways2. The assessment was performed using common cerebellar markers including NEPH3, PTF1A, OLIG2, and MATH1.
Based on the human brain in cerebellum—The Human Protein Atlas, 12 of high score (RS > 20) general cerebellum genes and 14 of specific regional cerebellar layer genes were assessed in this study. The expression of 10 out of the 12 new cerebellar markers was promoted in the RCP-derived cerebellar spheroids compared to the control. The expression of 13 out of the 14 makers for specific cerebellar layers was promoted in the RCP-derived cerebellar spheroids. These results further confirm the effects of RCP on the patterning of cerebellar spheroids from hiPSCs and provides 23 novel markers in characterization of early cerebellar differentiation. To further examine the response of RA, Wnt, and SHH activation, the downstream genes need to be investigated in future (Supplementary Table S9–11).
Moreover, our results show that the cerebellar marker-enriched RCP-derived spheroids exhibit more active glycolytic activity and anaerobic metabolism, which may provide important biomarkers for cerebellar differentiation and process monitoring. While the shift toward glycolysis pathway for the spheroids with stronger cerebellar identity may not be cerebellar-specific, the results may provide general metabolic insights in monitoring neural differentiation from hiPSCs44,45,46. Glucose is the major fuel source for energy metabolism under normal and malignant conditions for brain cells. Undifferentiated hPSCs mainly use glycolysis to maintain pluripotency and the early stage differentiation induces the shift towards OXPHOS10. However, different brain cell types and polarization status may induce the metabolic pathway shift47,48. The metabolic status corresponding to the cerebellar differentiation and maturation status may need further investigation.
Our results demonstrate the minimal effect of cerebellar phenotype on the EV biogenesis in the RCP-derived cerebellar spheroids. Most of the examined ESCRT-dependent and independent EV biogenesis markers are comparable at day 35 for RCP-derived cerebellar spheroids compared to the control. The few downregulated genes SMPD2 and HRS at day 35 were different from those downregulated at day 14. In general, the higher expression of EV biogenesis markers usually leads to the higher secretion of EVs. In this study, the differences in the EV mode size, EV numbers, and protein content were not statistically different around day 35, when the phenotype was mostly different for the control and RCP conditions. EV biogenesis could be affected by cytoskeleton alterations, hypoxia, pH, drug stimulation, and gene overexpression18,49. While the ability of the EV biogenesis of the RCP-derived cerebellar spheroids is retained, it is very likely that the protein and microRNA cargo of the secreted EVs is different due to the phenotypic difference of parent cells50,51. The EV cargo analysis usually requires the proteomics and microRNA profiling which has been shown for hiPSC-derived neural stem cells52.
Effect of Zinc on neural degeneration has been well recognized34,53. Histidine (His 6, His 13, and His 14) at the N-terminal domain of Aβ coordinates with Zn2+. Zinc is essential for brain function and tissue development. It is concentrated in the hippocampus and amygdala. Both Zn deficiency and Zn overload may affect cellular Zn homeostasis which is linked to neurodegeneration34,53. The predicted model for reactive oxygen species (ROS) level, cell viability, and cytotoxicity for zinc-induced toxicity in the derived cerebellar organoids is shown in Supplementary Fig. S3. Zn2+ ion is essential for cell development and scavenger protein production to reduce ROS level. Therefore, at low level of [Zn2+], it is hypothesized that cell viability increases until it reaches an optimal level; whereas, LDH and ROS levels decrease. When the Zn2+ level increases past the optimal concentration, it can induce ROS and harm the cells. Hence, cell viability may decrease fast and then slow down; whereas, LDH and ROS levels would increase fast and then are maintained at a certain value. Our results showed the mild zinc-induced neurotoxicity in the RCP-derived cerebellar spheroids when zinc exposure exceeds 1.0 µM. The effects of zinc might be different for different regions of human brain, which needs further investigation.
Conclusions
This study identified some early cerebellar markers and regionally enriched genes in the RCP-derived cerebellar spheroids from hiPSCs. The results also provide the characterizations of metabolic pathways showing the elevated glycolysis in the more mature cerebellar spheroids. The EV biogenesis ability was retained in the derived cerebellar spheroids with different phenotype. As a proof-of-concept study, mild zinc-induced neurotoxicity was observed in the RCP-derived cerebellar spheroids when zinc exposure exceeds 1.0 µM. This study should advance the understanding of biomarkers during early cerebellar development for cerebellum organoid engineering and neurotoxicity study.
Methods
Undifferentiated hiPSC culture
Human foreskin fibroblasts were transfected with plasmid DNA encoding reprogramming factors octamer-binding transcription factor 4 (OCT4), NANOG, SRY-box transcription factor 2 (SOX2), and LIN28 to produce human iPSK3 cells (kindly provided by Dr. Stephen Duncan, Medical College of Wisconsin)54,55. The hiPSCs were maintained in mTeSR Plus serum free medium (StemCell Technologies, Inc., Vancouver, Canada) on growth factor-reduced Geltrex-coated surface (Life Technologies). The cells were passaged every seven days using Accutase and seeded at 1 × 106 cells per well of six-well plate in the presence of rho-associated protein kinase (ROCK) inhibitor Y27632 (10 μM, Sigma) for the first 24 h56,57.
Cerebellar spheroid differentiation from hiPSCs
Cerebellar differentiation was modified based on the published protocol35,58 as reported in our previous study2. Human iPSK3 cells were seeded at 3 × 105 cells/well into ultra-low attachment 24-well plates (Corning Incorporated, Corning, NY) in differentiation medium composed of Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) with 2% B-27 serum-free supplements (Life Technologies). Y27632 (10 μM) and transforming growth factor (TGF)-β1 inhibitor SB431542 (10 μM, StemCell Technologies Inc.) were added for the first week of culture. On day 2, FGF2 (50 ng/mL, StemCell Technologies, Inc.) was added until the end of the first week. During the third week of culture, caudalization of neural rosettes in the derived aggregates was induced using FGF19 (100 ng/mL, Peprotech). Sequentially, the treatment of stromal cell-derived factor 1-α (SDF1A, 50 ng/mL, Peprotech) in the fifth week of culture was used to induce spheroid self-organization into the molecular layer, the Purkinje cell layer, and the granule cell layer. This procedure was used as a control condition.
The effects of casualization factor of retinoic acid (RA), wingless (Wnt) activator, and SHH activator were tested as shown in our previous study2. Briefly, during day 7–14, both RA (1.0 μM, Sigma) and Wnt activator CHIR99021 (CHIR, 10 μM, Sigma) were added to the spheroid culture of the control condition. In addition, during day 28–35, purmorphamine (PMR, 2 μM, Sigma) was used to activate the SHH pathway and promote ventralization of the spheroids. This condition was referred as RCP condition. On day 35, the gene and protein expression for cerebellar layer markers were characterized. As FGF8 has been reported to pattern rostral-caudal axis of during hPSC differentiation36, the effect of FGF8 treatment during day 7–21 was tested in comparison with RCP condition. For control and RCP groups, the spent media were collected and analyzed (n = 4) with a BioProfile Flex2 cell culture analyzer (Nova Biomedical) for metabolite analysis.
Flow cytometry
The spheroids were trypsinized and different neural marker expression levels were quantified. Then, 1 × 106 cells were fixed with 10% neutral formalin buffer (ThermoFisher) and washed with phosphate-buffered saline (PBS). The cells were permeabilized with 100% cold methanol for those with intracellular markers, blocked with blocking buffer (5% fetal bovine serum in PBS), and then incubated with different primary antibodies (Supplementary Table S1) followed by the corresponding secondary antibody Alexa Fluor 488 goat anti-Mouse IgG1. The cells were acquired using BD FACSCanto™ II flow cytometer (Becton Dickinson) and analyzed against isotype controls using FlowJo v10 software (https://www.bdbiosciences.com/en-us/products/software/flowjo-v10-software).
Immunocytochemistry
For biomarker detection, the cells were fixed using 4% paraformaldehyde (PFA) and permeabilized using 0.2% Trixton-X 100. The samples were blocked with 5% FBS in PBS and stained with the primary antibodies (Supplementary Table S1), followed by the corresponding anti-species Alexa Fluoro antibodies, i.e., Alexa Fluor 488 goat anti-mouse IgG1 or Alexa Fluor 594 goat anti-Rabbit IgG (Life technologies). Both primary and secondary antibody dilutions were made based on the manufacturer’s recommendations and prepared in staining buffer (2% FBS in PBS). Then the nuclei were counterstained with Hoechst 33342 (blue), and pictures were taken for blue, green, and red colors to detect the markers and their cellular locations under a fluorescent microscope (Olympus IX70, Melville, NY).
Extracellular vesicle isolation
Extracellular vesicles (EVs) were isolated from the conditioned media (around week 2–3 and week 5–6) by polyethylene glycol (PEG) precipitation and ultracentrifugation following our previous publications21,23. Briefly, cells were cultured in DMEM-F12 with serum-free B27 and the conditioned media were collected every 48–72 h. The conditioned media were then differential centrifuged (500g for 5 min; 2000g for 10 min; 10,000g for 30 min) to remove larger debris, apoptotic body and microvesicles. Supernatants were then mixed with PEG solution (24% wt/vol PEG, 1.5 M NaCl) at a 1:1 volume and incubated at 4 °C overnight. The next day, the mixed solutions were centrifuged at 3214g for 1 h to obtain crude EVs. The pellets were resuspended in PBS and then ultracentrifuged at 100,000g for 70 min. Purified EVs were resuspended in 100 µL particle-free PBS and stored in − 80 °C for further use.
Nanoparticle tracking analysis (NTA)
NTA was performed on the isolated EV samples in triplicate to determine size distribution and particle concentration. NTA was performed on a Nanosight LM10-HS instrument (Malvern Instruments, Malvern, UK) configured with a blue (488 nm) laser and CMOS camera59. For each replicate, three videos of 60 s were acquired with camera shutter speed fixed at 30.00 ms. To ensure accurate and consistent detection of small particles, camera level was set to 13, and detection threshold was maintained at 5. The laser chamber was cleaned thoroughly with particle-free water between each sample reading. The collected videos were analyzed using NTA3.4 software (NanoSight NTA software v3.4 in the instrument) to obtain the mode and mean size distribution, as well as the concentration of particles per mL of solution.
Reverse transcription-polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated from the neural spheroids using the RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol followed by the treatment of DNA-Free RNA Kit (Zymo, Irvine, CA). Reverse transcription was carried out using 2 μg of total RNA, anchored oligo-dT primers (Operon, Huntsville, AL), and Superscript III (Invitrogen, Carlsbad, CA) (according to the protocol of the manufacturer). Primers specific for target genes (Supplementary Table S2–S10) were designed using the Primer-BLAST (NCBI), and the melting temperature was checked using NetPrimer Analysis (PREMIER Biosoft). The gene β-actin was used as an endogenous control for normalization of expression levels. Real-time RT-PCR reactions were performed on an ABI7500 instrument (Applied Biosystems, Foster City, CA) using SYBR1 Green PCR Master Mix (Applied Biosystems). The amplification reactions were performed as following: 2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 95 °C for 15 s and 55 °C for 30 s, and 68 °C for 30 s. Fold variation in gene expression was quantified by means of the comparative 2−ΔΔCT method based on the comparison of expression of the target genes (normalized to the endogenous control β-actin) among different conditions.
Western blot assay
Cells were lysed in radio-immunoprecipitation assay (RIPA) buffer (150 mM sodium chloride, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris, pH 8, 2 µg/mL Aprotinin, 5 µg/mL Leupeptin, 5 µg/mL Antipain, 1 mM PMSF protease inhibitor), and homogenized by sonification using a Sonic Dismembrator 100 (Fisher Scientific, Hampton, NJ). Samples were then digested for 20 min on ice, and spun down at 14,000 rpm for 20 min. The supernatant was collected and a Bradford assay was carried out to determine the protein concentration. Protein lysate concentration was normalized, and 20 µg of each sample was denatured at 95 °C in 2 × Laemmli Sample buffer. Proteins were separated by 15% BIS–Tris-SDS gels and transferred onto a nitrocellulose membrane (Bio-rad, Hercules, CA). For the detection of non-phosphorylated proteins, the membranes were blocked for 30 min in 3% skim milk (w/v) in Tris-buffered saline (10 mM Tris–HCl, pH 7.5, and 150 mM NaCl) with 0.1% Tween 20 (v/v) (TBST), or in 3% bovine serum albumin in TBST. Membranes were incubated overnight in the presence of the primary antibody diluted in the corresponding blocking buffer at 4 °C. Afterward, the membranes were washed four times for 10 min each with TBST and then incubated with an IR secondary (LI-COR, Lincoln, NE) at 1:10,000 for 180 min at room temperature. Blots were washed another four times for 10 min each with TBST and processed using the LI-COR Odyssey (LI-COR). Images were analyzed using ImageJ 1.46r software (https://imagej.nih.gov/ij/, National Institutes of Health, USA) for band density, and the band density of proteins of interest was normalized to the band density of endogenous control β-actin.
Zinc-induced neurotoxicity
The derived day 35 cerebellar organoids were replated onto the Matrigel-coated 24-well plates and exposed to 0, 0.01, 0.1, 1.0, 10 and 100 µM Zinc supplemented culture media for three days. The supplementation of ZnSO4·7H2O (Sigma) to the media was performed to generate Zinc ion at the target concentrations. The cells were then examined by LIVE/DEAD assay and MTT assay. The spent media were saved for lactate dehydrogenase assay.
(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
The day 35 replated cerebellar spheroids exposed to different concentration of zinc were incubated with a 0.5 mg/mL MTT (Sigma) solution for an hour at 37 °C. The media and MTT were removed. The formazan crystals were dissolved in dimethyl sulfoxide and centrifuged at 800g for 5 min. The absorbance of the supernatants was measured at 490 nm on a microplate reader (BioRad Laboratories, Hercules, CA).
LIVE/DEAD staining
The cells were evaluated for viability using the LIVE/DEAD staining kit (Molecular Probes) according to the manufacturer’s protocol. The day 35 cerebellar spheroids were replated onto Matrigel-coated surface and exposed to Zinc at different concentrations. After Zinc treatment, the cells were washed with PBS and then incubated in DMEM-F12 containing 3–10 μM calcein-AM (green) and 8 μM ethidium homodimer I (red) for 20 min at room temperature and protected from light. The images were taken for the spheroid outgrowth under a fluorescent microscope (Olympus IX70, Melville, NY). Image analysis was performed using ImageJ 1.46r software (https://imagej.nih.gov/ij/, National Institutes of Health, USA) on at least three images for each condition. The viability was analyzed and calculated as the percentage of green intensity over total intensity (including both green cells and red cells).
Lactate dehydrogenase (LDH) activity assay
The cytotoxicity was assessed using LDH activity assay kit (Sigma, MAK066). Briefly, a total volume of 100 μL of spent medium of zinc-treated cerebellar spheroids and LDH reaction mixture were mixed. The initial absorbance at 450 nm was measured using a microplate reader (Bio-Rad iMark). The mixture was incubated at 37 °C and taken measurement every 5 min. The LDH activity was calculated through the subtraction of final and initial measurements in comparison to the standard curve.
Statistical analysis
The representative experiments were presented, and the results were expressed as [mean ± standard deviation]. To assess the statistical significance, one-way ANOVA or student’s t-test followed by Fisher’s LSD post hoc tests were performed. A p-value < 0.05 was considered statistically significant.
References
Roostaei, T., Nazeri, A., Sahraian, M. A. & Minagar, A. The human cerebellum: A review of physiologic neuroanatomy. Neurol. Clin. 32, 859–869 (2014).
Hua, T. et al. Cerebellar differentiation from human stem cells through Retinoid, Wnt, and sonic hedgehog pathways. Tissue Eng. Part A 27, 881–893 (2021).
Aldinger, K. A. et al. Spatial and cell type transcriptional landscape of human cerebellar development. Nat. Neurosci. 24, 1163–1175 (2021).
Suzuki, I. K. & Vanderhaeghen, P. Is this a brain which I see before me? Modeling human neural development with pluripotent stem cells. Development 142, 3138–3150 (2015).
Tao, Y. & Zhang, S. C. Neural subtype specification from human pluripotent stem cells. Cell Stem Cell 19, 573–586 (2016).
Bradley, R.A. et al. Regionally specified human pluripotent stem cell-derived astrocytes exhibit different molecular signatures and functional properties. Development 146, dev170910 (2019).
Rifes, P. et al. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat. Biotechnol. 6, 66 (2020).
Muguruma, K. et al. Ontogeny-recapitulating generation and tissue integration of ES cell-derived Purkinje cells. Nat. Neurosci. 13, 1171–1180 (2010).
Silva, T. P. et al. Maturation of human pluripotent stem cell-derived cerebellar neurons in the absence of co-culture. Front. Bioeng Biotechnol. 8, 70 (2020).
Sart, S., Agathos, S. N. & Li, Y. Process engineering of stem cell metabolism for large scale expansion and differentiation in bioreactors. Biochem. Eng. J. 84, 74–82 (2014).
Yuan, X. et al. NAD+/NADH Redox alterations reconfigure metabolism and rejuvenate senescent human mesenchymal stem cells in vitro. Commun. Biol. 3, 774 (2020).
Jeske, R. et al. In vitro culture expansion shifts the immune phenotype of human adipose-derived mesenchymal stem cells. Front. Immunol. 12, 621744 (2021).
Tanosaki, S., Tohyama, S., Kishino, Y., Fujita, J. & Fukuda, K. Metabolism of human pluripotent stem cells and differentiated cells for regenerative therapy: a focus on cardiomyocytes. Inflamm. Regen. 41, 5 (2021).
Moussaieff, A. et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 21, 392–402 (2015).
O’Brien, L. C., Keeney, P. M. & Bennett, J. P. Jr. Differentiation of human neural stem cells into motor neurons stimulates mitochondrial biogenesis and decreases glycolytic flux. Stem Cells Dev. 24, 1984–1994 (2015).
Colombo, M., Raposo, G. & Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 (2014).
Riazifar, M., Pone, E. J., Lotvall, J. & Zhao, W. Stem cell extracellular vesicles: Extended messages of regeneration. Annu. Rev. Pharmacol. Toxicol. 57, 125–154 (2017).
Liu, C. & Su, C. Design strategies and application progress of therapeutic exosomes. Theranostics 9, 1015–1028 (2019).
Zhang, X. et al. Engineered extracellular vesicles for cancer therapy. Adv. Mater. 33, e2005709 (2021).
Colao, I. L., Corteling, R., Bracewell, D. & Wall, I. Manufacturing exosomes: A promising therapeutic platform. Trends Mol. Med. 24, 242–256 (2018).
Marzano, M. et al. Differential effects of extracellular vesicles of lineage-specific human pluripotent stem cells on cellular behaviours of isogenic cortical spheroids. Cells 8, 993–1014 (2019).
Jeske, R., Bejoy, J., Marzano, M. & Li, Y. Human induced pluripotent stem cell-derived extracellular vesicles: Characteristics and applications. Tissue Eng. Part B Rev. 26, 129–144 (2020).
Marzano, M. et al. Biogenesis of extracellular vesicles produced from human stem cell-derived cortical spheroids exposed to iron oxides. ACS Biomater. Sci. Eng. 7, 1111–1122 (2021).
Andersen, J. et al. Generation of functional human 3D Cortico-motor assembloids. Cell 183, 1913–1929e1926 (2020).
Song, L. et al. Assembly of human stem cell-derived cortical spheroids and vascular spheroids to model 3-D brain-like tissues. Sci. Rep. 9, 5977 (2019).
Garreta, E. et al. Rethinking organoid technology through bioengineering. Nat Mater 20, 145–155 (2021).
Del Dosso, A., Urenda, J. P., Nguyen, T. & Quadrato, G. Upgrading the physiological relevance of human brain organoids. Neuron 107, 1014–1028 (2020).
Song, L. et al. Functionalization of brain region-specific spheroids with isogenic Microglia-like cells. Sci. Rep. 9, 11055–11072 (2019).
Wilhelm, I., Nyul-Toth, A., Suciu, M., Hermenean, A. & Krizbai, I. A. Heterogeneity of the blood–brain barrier. Tissue Barriers 4, e1143544 (2016).
Tan, H. Y., Cho, H. & Lee, L. P. Human mini-brain models. Nat. Biomed. Eng 5, 11–25 (2021).
Zieminska, E., Ruszczynska, A. & Lazarewicz, J. W. Tetrabromobisphenol A disturbs zinc homeostasis in cultured cerebellar granule cells: A dual role in neurotoxicity. Food Chem. Toxicol. 109, 363–375 (2017).
Mezzaroba, L., Alfieri, D. F., Colado Simao, A. N. & Vissoci Reiche, E. M. The role of zinc, copper, manganese and iron in neurodegenerative diseases. Neurotoxicology 74, 230–241 (2019).
Chiaradia, I. & Lancaster, M. A. Brain organoids for the study of human neurobiology at the interface of in vitro and in vivo. Nat. Neurosci. 23, 1496–1508 (2020).
Miller, Y., Ma, B. & Nussinov, R. Zinc ions promote Alzheimer Abeta aggregation via population shift of polymorphic states. Proc. Natl. Acad. Sci. USA 107, 9490–9495 (2010).
Muguruma, K., Nishiyama, A., Kawakami, H., Hashimoto, K. & Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 10, 537–550 (2015).
Imaizumi, K. et al. Rostrocaudal areal patterning of human PSC-derived cortical neurons by FGF8 signaling. eNeuro 5, 66 (2018).
The human brain in cerebellum—The Human Protein Atlas. http://www.proteinatlas.org/humanproteome/brain/cerebellum (2003).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Colombo, M. et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126, 5553–5565 (2013).
Silva, T. P. et al. Scalable generation of mature cerebellar organoids from human pluripotent stem cells and characterization by immunostaining. J. Vis. Exp. 6, 66 (2020).
Fleck, J.S. et al. Resolving organoid brain region identities by mapping single-cell genomic data to reference atlases. Cell Stem Cell 28, 1148–1159e1148 (2021).
Valiulahi, P. et al. Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs. Stem Cell Rep. 6, 66 (2021).
Libby, A. R. G. et al. Axial elongation of caudalized human organoids mimics aspects of neural tube development. Development 148, 66 (2021).
Vacanti, N. M. & Metallo, C. M. Exploring metabolic pathways that contribute to the stem cell phenotype. Biochim Biophys, Acta 1830, 2361–2369 (2013).
Sa, J. V. et al. Quantification of metabolic rearrangements during neural stem cells differentiation into astrocytes by metabolic flux analysis. Neurochem. Res. 42, 244–253 (2017).
Perestrelo, T., Correia, M., Ramalho-Santos, J. & Wirtz, D. Metabolic and mechanical cues regulating pluripotent stem cell fate. Trends Cell Biol. 28, 1014–1029 (2018).
Orihuela, R., McPherson, C. A. & Harry, G. J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 173, 649–665 (2016).
Nwadozi, E., Rudnicki, M. & Haas, T. L. Metabolic coordination of pericyte phenotypes: Therapeutic implications. Front. Cell Dev. Biol. 8, 77 (2020).
Patel, D. B., Santoro, M., Born, L. J., Fisher, J. P. & Jay, S. M. Towards rationally designed biomanufacturing of therapeutic extracellular vesicles: Impact of the bioproduction microenvironment. Biotechnol. Adv. 36, 2051–2059 (2018).
Zaborowski, M. P., Balaj, L., Breakefield, X. O. & Lai, C. P. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience 65, 783–797 (2015).
Jeppesen, D.K. et al. Reassessment of exosome composition. Cell 177, 428–445e418 (2019).
Upadhya, R. et al. Extracellular vesicles from human iPSC-derived neural stem cells: miRNA and protein signatures, and anti-inflammatory and neurogenic properties. J. Extracell Vesicles 9, 1809064 (2020).
Yuan, Y., Niu, F., Liu, Y. & Lu, N. Zinc and its effects on oxidative stress in Alzheimer’s disease. Neurol. Sci. 35, 923–928 (2014).
Si-Tayeb, K. et al. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev. Biol. 10, 81 (2010).
Si-Tayeb, K. et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297–305 (2010).
Song, L., Wang, K., Li, Y. & Yang, Y. Nanotopography promoted neuronal differentiation of human induced pluripotent stem cells. Colloids Surf. B 148, 49–58 (2016).
Bejoy, J., Song, L., Zhou, Y. & Li, Y. Wnt-Yes associated protein interactions during neural tissue patterning of human induced pluripotent stem cells. Tissue Eng. Part A 24, 546–558 (2018).
Muguruma, K. Self-organized cerebellar tissue from human pluripotent stem cells and disease modeling with patient-derived iPSCs. Cerebellum 17, 37–41 (2018).
Rider, M. A., Hurwitz, S. N. & Meckes, D. G. Jr. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 6, 23978 (2016).
Acknowledgements
The authors thank Flow Cytometry Laboratory of FSU Department of Biomedical Sciences, Dr. Brian K. Washburn and Kristina Poduch from the Molecular Cloning Facility of FSU Department of Biological Sciences for their help with RT-PCR analysis, Dr. Stephen Duncan of Medical College of Wisconsin and Dr. David Gilbert of FSU Department of Biological Sciences for human iPSK3 cells. This work is partially supported by NSF Career Award 1652992 (to YL), NSF 1917618 (to YL), and Florida Department of Health (FDOH) Live Like Bella Award (9LA01 to QXS and YL).
Author information
Authors and Affiliations
Contributions
T.H. and C.L. performed most of experiments and wrote the main manuscript. S.K. helped on the experiments. K.G., and S.J. performed metabolite analysis. D.M. helped extracellular vesicle isolation and characterization. Y.L. and Q.S. conceived the experiments, wrote, and finalized the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hua, T., Liu, C., Kiran, S. et al. Phenotypic, metabolic, and biogenesis properties of human stem cell-derived cerebellar spheroids. Sci Rep 12, 12880 (2022). https://doi.org/10.1038/s41598-022-16970-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-16970-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.