Abstract
Trans-2,3-dihydro-3-hydroxyanthranilic acid (DHHA) is a cyclic β-amino acid used for the synthesis of non-natural peptides and chiral materials. And it is an intermediate product of phenazine production in Pseudomonas spp. Lzh-T5 is a P. chlororaphis strain isolated from tomato rhizosphere found in China. It can synthesize three antifungal phenazine compounds. Disruption the phzF gene of P. chlororaphis Lzh-T5 results in DHHA accumulation. Several strategies were used to improve production of DHHA: enhancing the shikimate pathway by overexpression, knocking out negative regulatory genes, and adding metal ions to the medium. In this study, three regulatory genes (psrA, pykF, and rpeA) were disrupted in the genome of P. chlororaphis Lzh-T5, yielding 5.52 g/L of DHHA. When six key genes selected from the shikimate, pentose phosphate, and gluconeogenesis pathways were overexpressed, the yield of DHHA increased to 7.89 g/L. Lastly, a different concentration of Fe3+ was added to the medium for DHHA fermentation. This genetically engineered strain increased the DHHA production to 10.45 g/L. According to our result, P. chlororaphis Lzh-T5 could be modified as a microbial factory to produce DHHA. This study laid a good foundation for the future industrial production and application of DHHA.
Similar content being viewed by others
Introduction
Trans 2,3-dihydro-3-hydroxyanthranilic acid (DHHA) can be used in cycloaddition reactions as an enantiomerically pure building block, for biosynthesis of unnatural peptides, and for preparation of various useful intermediate acid derivatives of benzoic acid, such as 3-hydroxyanthranilic acid and anthranilic acid, which are important aromatic compounds1,2,3,4. These compounds are widely used in chemicals, food, cosmetics, and pharmaceuticals. In the current market, production of aromatic compounds relies heavily on direct extraction from plants or petroleum-derived chemical processes. The demand for establishing new sustainable sources and renewable aromatics has increased rapidly in recent years. The sustainable production of aromatics has drawn great interest5,6,7,8. Microbial bioproduction using abundant feedstocks is a highly promising alternative8.
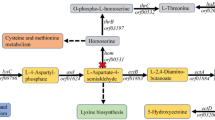
In recent years, the use of renewable resources to produce chemicals and fuels has attracted the attention of researchers9,10,11. Different than other aromatic compounds, DHHA can be synthesized in microorganisms. McCormick et al. first isolated DHHA from the fermentation broth of Streptomyces aureofaciens S-652 and Meade et al. obtained an S. aureofaciens strain which had a production of 8 g/L of DHHA after 120 h of fermentation3,12. According to previous research by Mavrodi et al., benzoate was converted to 2-amino-4-deoxycholic acid (ADIC) by PhzE, and then converted by PhzD to DHHA, in certain Pseudomonas strains. DHHA is an important intermediate of the phenazine biosynthesis of Pseudomonas spp (Fig. 1)13,14.
Central carbon metabolism related to the biosynthesis of phenazines in P. chroloraphis Lzh-T5. The enzymes: pgi, phosphoglucose isomerase; glk, glucokinase; eno, enolase; gapA, glyceraldehyde 3-phosphate dehydrogenase; glpK, glycerol kinase; glpF, glycerol facilitator; fda, fructose-1,6-P2 aldolase; glpD, glycerol-3-P dehydrogenase; fbp, fructose 1,6-bisphosphatase; tpiA, triosephosphate isomerase; talB, transaldolase; zwf, G6P dehydrogenase; pck, PEP carboxykinase; ppc, PEP carboxylase; pgm, phosphoglyceromutase; pgk, phosphoglycerate kinase; phzF, asparagine synthase. DHAP dihydroxyacetone phosphate; Gly3P Glycerol 3-phosphate; G6P glucose 6-phosphate; F16BP fructose 1,6-bisphosphate; GAP glyceraldehyde 3-phosphate; F6P fructose 6-phosphate; 6PGNL 6-phosphogluconolactone; R5P ribose 5-phosphate; Ru5P ribulose 5-phosphate; S7P sedoheptulose 7-phosphate; Xu5P xylulose 5-phosphate; PEP phosphoenolpyruvate; E4P erythrose 4-phosphate; ACoA acetyl-coenzyme A; PYR pyruvate; OAA oxaloacetate; CIT citrate; DHQ 3-dehydroquinic acid; DAHP 3-deoxy-Darabinoheptulosonate7-phosphate; QA quinic acid; DHS 3-dehydroshikimic acid; SA shikimic acid; GA gallic acid; CHO chorismate.
Lzh-T5 is a P. chlororaphis strain isolated from the tomato rhizosphere located in China. It has a phenazine biosynthesis cluster phzABCDEFG, and could produce phenazine-1-carboxylic acid (PCA) and other phenazine derivatives15. In this study, phzF of P. chlororaphis Lzh-T5 was disrupted causing DHHA accumulation. Three negative regulatory genes (pykF, psrA, and rpeA) were stepwise disrupted in P. chlororaphis Lzh-T5. The yield of DHHA increased from 2.15g/L to 5.52 g/L. To improve DHHA production, key genes were overexpressed by BglBrick vectors from the shikimate, pentose phosphate, and gluconeogenesis pathways of Lzh-T5, obtaining 7.89 g/L DHHA. The effect of Fe3+ on DHHA production was investigated. A final DHHA yield of 10.45 g/L was obtained.
Methods
Microorganisms, growth conditions, and plasmids
All plasmids, primers, and microorganisms are listed in Table 1 and Supplementary Material 1: Table S1. All Escherichia coli strains were cultured in LB medium at 37 °C, and all P. chlororaphis strains were cultured in KB medium at 30 °C. Kanamycin and Ampicillin were used. All details are detailed in our previous research16.
DNA manipulation
A no-scar deletion method was used in the genome of P. chlororaphis Lzh-T5 and its derivative strain. In order to create LDA-1 by the interruption of phzF in Lzh-T5, two pairs of primers (phzF-A (EcoRI)–phzF-B and phzF-C–phzF-D (XbaI)) were designed. Upstream (740 bp) and downstream (793 bp) of phzF were first amplified by PCR. A 1515 bp fragment fusion was amplified by overlap PCR. The fusion fragment was constructed into pK18mobsacB, creating the recombinant plasmid pK18-phzF.
The pK18-phzF plasmid was transferred to E. coli S17-1 (λpir) by heat shock transformation. Then biparental mating between E. coli S17-1 and P. chlororaphis Lzh-T5 generated the mutant LDA-1 strain. Single-crossover and double-crossover clones were selected stepwise. To ensure accuracy, PCR analysis and sequencing were used to confirm the deletion. Detailed steps are listed in our previous research16. Similarly, rpeA, pykF, and psrA were disrupted in their corresponding strains.
BglBrick plasmids are a kind of widely used Brick plasmids17. The plasmid pBbB5K-GFP was used as the backbone to overexpress six key genes. Recombinant plasmids used in this study were constructed following the methods described in our previous research16. In brief, six Brick plasmids containing aroB, aroD, aroE, phzC, tktA, and ppsA, respectively, were first constructed. Point mutations were made in order to remove restriction sites (EcoRI, HindIII, BamHI, and BglII) in aroD, tktA, and ppsA. Then, a complicated overexpression plasmid pBbB5K-aroE-aroD-aroB-phzC-tktA-ppsA was constructed following the BglBrick standard.
Quantitative RT-PCR
Quantitative RT-PCR was used to detect the transcriptional changes of related genes of different strains. And the rpoD which is one of housekeeping gene in P. chlororaphis was selected as the internal reference gene. The measurement methods of Quantitative RT-PCR are following our previous work18. And the fold change for mRNAs was calculated by the 2−ΔΔCt method19.
Fermentation processing
All P. chlororaphis strains were stored in an ultra-low temperature refrigerator. Strains were activated in KB petriplate with Ampicillin at 30 °C for 12–24 h before fermentation. Single colonies were inoculated in a 50 mL flask which contains 5 mL of KB medium and incubated overnight. Portions of seed bacteria were then inoculated in 50 mL KB medium in a 250 mL baffled flask and inoculation initial OD600 was 0.03. The gene expression of BglBrick plasmids was induced by isopropyl-β-d-thiogalactopyranoside (IPTG). 50 μM IPTG was used during an incubation of 12 h. After growing at 30 °C and centrifuged at 200 rpm for 24–72 h, the fermentation broth was collected to measure phenazine compounds and OD600. All experiments were performed in triplicate, and data were averaged and reported as mean ± standard deviation. After growing at 30 °C and 200 centrifuged at rpm for 24–72 h, the culture was collected for measurement of phenazine compounds and OD600. All fermentation experiments were performed in triplicate, and the data were averaged and reported as mean ± standard deviation.
Measurement, purification, and quantification of DHHA from fermentation broth
Determination and purification of DHHA from fermentation broth followed the methods described in previous research20. Briefly, the supernatant of the fermentation broth was collected by centrifuging at 12,000g for 15 min. Then, it was analyzed by liquid chromatography–mass spectrometry (LC–MS) after processing through a 0.22 μm polyvinylidene difluoride syringe filter. LC–MS was performed on the Agilent HPLC1290-MS6230 system (Agilent Technologies, Santa Clara, CA) by an Agilent Extend C18 column (50 mm × 2.1 mm, 1.8 μm). It was eluted with methanol/0.1% formic acid (50:50, v/v) at a 0.15 mL/min flow rate. The DHHA sample was analyzed by mass spectrometry in the positive ion detection mode.
In order to purify DHHA, Shimadzu Inert Sustain phenyl column (20 × 250 mm, 15 μm) was used in HPLC (Shimadzu LC8A, Shimadzu, Kyoto, Japan). It was eluted with water/methanol (90:10, v/v) with a flow rate of 2 mL/min at a scanning wavelength of 278 nm. We collected the peak containing DHHA and dried it by vacuum freezing. The crystals of DHHA were obtained at room temperature after dissolving them in hot ethanol (65 °C).
Quantification of DHHA was described in our previous work 18. Briefly, the fermentation broth was centrifuged at 11,000 rpm for 3 min and supernatant collected. The samples were analysis by HPLC (Agilent 1260, USA) with a Shimadzu Inert Sustain phenyl column (4.6 × 250 mm, 5 μm) to determine the amount of DHHA. It was eluted with 0.1% formic acid/methanol (85:15, v/v) with a flow rate of 1 mL/min at a scanning wavelength of 278 nm. A DHHA standard curve was used to infer the DHHA content by plotting the concentration of the DHHA standard solution on the abscissa and the corresponding absorbance (peak area) on the vertical.
Superoxide dismutase activity measurement
In the Fe3+ addition experiments, superoxide dismutase (SOD) activities of different strains were measured. The methods of measure are following the previous literature21,22.
Results
Disruption of phenazine synthesis in Lzh-T5
According to the research from Blankenfeldt, DHHA undergoes an isomerization reaction facilitated by PhzF and converts to 6-amino-5-oxocyclohex-2-ene-1-carboxylic acid in Pseudomonas spp23. In order to accumulate DHHA, phzF was disrupted in P. chlororaphis Lzh-T5, and the strain P. chlororaphis LDA-1 was got. As shown in Fig. 2, after 48 h of culture, the P. chlororaphis LDA-1 strain on an agar plate turns milky; however, the colony color of Lzh-T5 remains orange. This suggests that the phenazine derivatives could not be synthesized in LDA-1. After fermentation and analysis by HPLC–UV, phenazines (including 2-hydroxyphenazine, PCA, and 2-OH-PCA) disappeared in the broth of P. chlororaphis LDA-1 (Fig. 3).
Similar to previous research, a new absorption peak appears in the HPLC–UV at a wavelength of 278 nm, which is the maximum absorption wavelength of DHHA (Fig. S1)20. According to the analysis of LC–MS, the mass-to-charge ratio (m/z) of the compound was 156.0658 for [C7H9NO3+ H]+ (the mass of DHHA is 155.0655; Fig. S1). When phzF is overexpressed in the LDA-1 strain, production of phenazine was recovered (Fig. 3). According to these results, it suggests that disruption of phzF in Lzh-T5 causes accumulation of DHHA. Using HPLC–UV analysis, the DHHA yield of LDA-1 reached 2.15 g/L in 48 h (Fig. 4a). Our results suggest that disruption of phzF had little effect on Lzh-T5 cell growth (Fig. 4b).
Knockout of negative regulatory genes to boost DHHA production
According to previous research, interruption the pykF (which encode pyruvate kinase) enhances the production of 2-hydroxyphenazine16. In this study, pykF from LDA-1 was initially chosen to inactivate, so we obtained a mutant strain LDA-2 (Fig. 5). After analysis of the fermentation broth by HPLC–UV, the DHHA yield of strain LDA-2 increased from 2.15 g/L to 4.17 g/L (Fig. 5a). The growth condition of the strain was detected to have little effect after deletion of pykF (Fig. 5b).
PsrA, a sigma regulator, was first reported in P. chlororaphis PCL139124. According to Chin-A-Woeng et al., PsrA played a negative regulatory role in the production of the antifungal metabolite PCA in P. chlororaphis PCL139124. Similar results were obtained from psrA disruption in P. chlororaphis HT66 (Peng et al. 2018)25. According to our research, the sigma regulator psrA also exists in P. chlororaphis Lzh-T5. The strain LDA-3 was obtained after the gene psrA was disrupted in LDA-2, and the production of DHHA increased from 4.17 g/L to 4.92 g/L (Fig. 5a).
RpeA, a negative regulator of Phenazine, was mutated by insertion in P. chlororaphis GP72 and results in a production increase of 2-hydroxyphenazine16,26. RpeA is part of the two-component signal transduction system (TCST) RpeA/RpeB, and is present in other Pseudomonas strains. For example, in P. chlororaphis 30–84, an RpeA homologue, negatively regulates the yield of PCA, indicating a conserved mechanism of Pseudomonas spp in phenazine synthesis regulation27,28. In this study, rpeA was disrupted in the LDA-3 genome to construct LDA-4. Similar to the insertional mutagenesis of P. chlororaphis GP72, DHHA yield of LDA-4 increased from 4.92 g/L to 5.52 g/L (Fig. 5a). After knocking out the negative regulatory genes, quantitative RT-PCR results showed that the transcript level of genes phzD and phzE in the derivative strains, which is key gene for DHHA synthesis, has increased significantly (Fig. S2).
Enhanced DHHA production by key gene overexpression
We disrupted pykF to improve the yield of DHHA from 2.15 g/L to 4.17 g/L by diverting more metabolic flux into the shikimate pathway from other pathways (Fig. 5a). This indicates that enhancing the lead synthesis pathway, we could improve the yield of DHHA in P. chlororaphis Lzh-T5. Compared with knocking out negative regulatory genes, gene overexpression is another effective strategy often used to increase the yield of biologic products in microorganisms. According to previous research, overexpression of key genes in the shikimate pathway enhanced the yield of 2-OH-PHZ16. In order to increase the production of DHHA, aroB, aroD, aroE, phzC, tktA, and ppsA from Lzh-T5 were overexpressed from the shikimate, pentose phosphate, and gluconeogenesis pathways. We used a previously employed kind of modular vector, the BglBrick plasmid29. A recombinant plasmid containing six genes, pBbB5K-aroE-aroD-aroB-phzC-tktA-ppsA, was constructed. Strain LDA-5 was created after transformation into the strain LDA-4 by electrotransformation. After fermentation, the DHHA production of LDA-5 increased to 7.89 g/L after 48 h (Fig. 5a). This indicates that overexpression of key genes is an effective strategy to enhance the production of DHHA. Quantitative RT-PCR results showed that the transcript level of six genes overexpressed in the derivative strain LDA-5 has increased (Fig. S3).
Enhanced DHHA production with Fe3+
Environmental factors have important effects on secondary metabolite production in Pseudomonas strains, especial ion concentration in the medium30,31. There is no universal medium suitable for Pseudomonas strains which can produce phenazines due to different nutritional requirements30. According to previous research, the DHHA production has a 30% increase after adding 3 mM of Fe3+20. To improve the production of DHHA, the effect of different concentrations of Fe3+ in the medium was investigated. After fermentation, DHHA production was detected by HPLC–UV. Low concentration of iron ions promoted DHHA production. High concentrations of iron ions inhibited DHHA production (Fig. 6). Different from our previous research, in the fermentation of LDA-5, the optimum concentration which enhanced the production of DHHA was 2 mM. We obtained a maximum DHHA yield of 10.45 g/L with 2 mM Fe3+ (Fig. 6a).
Discussion
Pseudomonas spp is a class of microorganism that exists widely in the environment32. It has strong adaptability and often produces resistant substances. Among these substances, phenazine derivatives are typical secondary metabolites, such as PCA, Pyocyanin (PYO), 2-Hydroxyphenazine and phenazine-1-carboxamide (PCN)25,33. Phenazine derivatives are produced by the phz gene clusters (phzABCDEFG) found in several Pseudomonas spp (including P. chlororaphis, P. fluorescence, and P. aeruginosa)34,35,36. In P. chlororaphis, phzC catalyzes the first reaction of the shikimic acid pathway, by catalysis of E4P and PEP to synthesize DAHP13. PhzE, the first key enzyme in the phenazine synthesis pathway which catalyzes the last product of the shikimate pathway into 2-amino-4-deoxy branched acid37. ADIC converts to DHHA and pyruvate by the isobranse enzyme PhzD23. It is then epimerized with diaminopimelate enzyme (diaminopimelate epimerase (DapF)) PhzF to 6-amino-5-oxocyclohex-2-ene-1-carboxylic acid (AOCHC)38. Two molecules of AOCHC are converted to hexahydrophenazine-1,6-dicarboxylic acid (HHPDC) catalyzed by the dimer PhzAB23. HHPDC spontaneously undergoes oxidative decarboxylation to form tetrahydrophenazine-1-carboxylic acid (THPCA). THPCA is catalyzed by PhzG to form 5,10-dihydro-phenazine-1-carboxylic acid (DHCCA)39, DHCCA eventually undergoes self-oxidation in the air to form PCA. While DHHA is an important intermediate product of phenazines, the production of phenazines is quite low. For example, in wild type P. chlororaphis GP72, the yield of 2-OH-PHZ is 4.5 mg/L, and the higher phenazine (PCN) production in wild type is about 400 mg/L in P. chlororaphis HT6625,26. The phenazine production strains are also the potential DHHA producers. P. aeruginosa is an opportunistic pathogenic bacterium, while P. chlororaphis is not. P. chlororaphis was selected as a candidate for DHHA production16. According to previous research, P. chlororaphis GP72 could accumulate 1.92 g/L DHHA with phzF disruption20. Lzh-T5 is a P. chlororaphis strain isolated from the tomato rhizosphere found in China. It has the phenazine biosynthesis cluster phzABCDEFG, and can produce phenazine derivatives15. In this study, phzF was disrupted from the genome of P. chlororaphis Lzh-T5 making the strain P. chlororaphis LDA-1 which has 2.15 g/L DHHA accumulation.
Enhancing the shikimate pathway is an effective strategy for aromatic compound production in microorganisms20. The direct precursors of the shikimate pathway are PEP and E4P. The PEP and E4P supply could be enhanced by an overexpression of PEP synthase encoded by ppsA and transketolase encoded by tktA1,40. In addition, inactive pykF, which encodes pyruvate kinase, can increase PEP41,42. Further carbon flux of the shikimate pathway impedes enzymatic reactions and removes the allosteric and transcriptional regions20,43. Quinate/shikimate dehydrogenase, dehydroquinic acid synthase, dehydroquinic acid dehydratase, and DHAP synthetase, encoded by aroE, aroB, aroD, and phzC, respectively, were reported as limiting steps in the shikimate pathway44. Our previous studies showed that disruption of pyruvate kinase (encoded by pykF) and coexpression of ppsA, tktA, aroB, aroD, aroE, and phzC in GP72 increased phenazine production20. In this study, the disruption of pykF and six gene tktA, ppsA, aroB, aroD, aroE, and phzC coexpression in LDA-1 increased the DHHA production to 7.89 g/L.
TCST systems exist in Pseudomonas spp that help them adapt to the environment by coordinating cellular pathways to interact with the environment28. Different TCST systems were found in P. chlororaphis Lzh-T5. Among these TCST systems, GacS/GacA is one of the most researched. GacS/GacA was the earliest TCST system which could enhance bioactive secondary metabolite production in Pseudomonas spp45,46. In P. chlororaphis HT66, GacA positively regulates the expression of psrA. psrA negatively controls the expression of rpoS and the expression of phenazine25. In this study, psrA was found in P. chlororaphis Lzh-T5. Interruption of psrA increased the production of DHHA from 4.17 g/L to 4.92 g/L, this suggests that psrA negatively controls the production of DHHA. RpeA/RpeB is a TCST system found widely in Pseudomonas spp28. According to the research of Whistler et al., Pip improved the production of PCA by enhancing the expression of phzR and phzI. Sigma factor rpoS regulated the expression of pip, which itself is regulated by the RpeA/RpeB TCST system. RpeB was inhibited by RpeA, so an rpeA mutant strain enhanced the production of phenazine28. Similar results were observed from P. chlororaphis GP72 and P. chlororaphis HT6616,25. The rpeA/rpeB TCST system was found in P. chlororaphis Lzh-T5, interruption of rpeA results in the increase in DHHA from 4.92 g/L to 5.52 g/L (Fig. 5a). This result suggests that, similar with P. chlororaphis 30–84, rpeA negatively impacts the production of DHHA.
Metal ions are an important factor which could affect the production of secondary metabolites of Pseudomonas spp30. Fe3 + is one of the important ions, and according to the research of Shtark et al., it plays a positive role in the production of phenazines in P. chlororaphis SPB1217. They hypothesize that Fe3+ activates some dependent superoxide dismutases and those superoxide dismutases promotes phenazine production by removing reactive oxygen species or bacterial metabolites that suppress enzymes involved in the synthesis pathway of phenazines30. Similar results were reported in P. aeruginosa PCL1391 and P. fluorescens 2–7947,48. Fe3+ also had a positive effect on DHHA production20. And our results shown that the SOD activity of the strain has increased after Fe3+ adding to the medium (Fig. S4). In this study, adding different concentration of Fe3+ had different effects on DHHA production in P. chlororaphis LDA-5. Low concentrations of Fe3+ promoted DHHA production, while high concentrations of Fe3+ inhibited DHHA production (Fig. 6). Adding 2 mM of Fe3+ had a positive effect on DHHA production, and our maximum DHHA yield was 10.45 g/L in this study.
In conclusion, phzF of P. chlororaphis Lzh-T5 was disrupted to construct the strain P. chlororaphis LDA-1 which had DHHA accumulation. Then, three negative regulatory genes (pykF, psrA and rpeA) were disrupted stepwise in P. chlororaphis LDA-1. The production of DHHA increased from 2.15 g/L to 5.52 g/L. Next, different key genes selected from the shikimate, pentose phosphate, and gluconeogenesis pathways of Lzh-T5, were overexpressed by BglBrick vectors. The production of DHHA increased from 5.52 g/L to 7.89 g/L. The effect of adding Fe3+ on DHHA production was investigated in a strain (LDA-5) with a resulting DHHA yield of 10.45 g/L.
Data availability
All data generated or analyzed during this study are included in this published article (and its Additional file).
References
Juaristi, E. & Soloshonok, V. Enantioselective Synthesis of Beta-amino Acids (Wiley, 2005).
Bunnage, M. E., Ganesh, T., Masesane, I. B., Orton, D. & Steel, P. G. Asymmetric synthesis of the putative structure of (−)-oryzoxymycin. Org. Lett. 5(3), 239–242 (2003).
Meade, T.J. Synthesis of aromatic heterocyclic polymers from a biosynthetically prepared precursor. US Patent. 5,340,913 (1994).
Palko, M., Kiss, L. & Fulop, F. Syntheses of hydroxylated cyclic beta-amino acid derivatives. Curr. Med. Chem. 12(26), 3063–3083 (2005).
Lee, S. Y. et al. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2(1), 18–33 (2019).
Keasling, J. D. Manufacturing molecules through metabolic engineering. Science 330(6009), 1355–1358 (2010).
Nielsen, J. Metabolic engineering. Appl. Microbiol. Biotechnol. 55(3), 263–283 (2001).
Liu, Q. et al. Current state of aromatics production using yeast: Achievements and challenges. Curr. Opin. Biotechnol. 65, 65–74 (2020).
Becker, J. et al. Top value platform chemicals: Bio-based production of organic acids. Curr. Opin. Biotechnol. 36, 168–175 (2015).
Choi, S. et al. Biorefineries for the production of top building block chemicals and their derivatives. Metab. Eng. 28, 223–239 (2015).
Noda, S. & Kondo, A. Recent advances in microbial production of aromatic chemicals and derivatives. Trends Biotechnol. 35(8), 785–796 (2017).
McCormick, J. et al. (+) trans-2,3-dihydro-3-hydroxyanthranilic acid. A new amino acid produced by Streptomyces aureofaciens. J. Am. Chem. Soc. 83, 4104–4105 (1961).
Mavrodi, D. V. et al. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2–79. J. Bacteriol. 180(9), 2541–2548 (1998).
McDonald, M. et al. Phenazine biosynthesis in Pseudomonas fluorescens: Branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. J. Am. Chem. Soc. 123(38), 9459–9460 (2001).
Li, Z. et al. Genome sequence of Pseudomonas chlororaphis Lzh-T5, a plant growth-promoting rhizobacterium with antimicrobial activity. Genome Announc. 6(18), e00328 (2018).
Liu, K. et al. Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-Hydroxyphenazine. Microb. Cell Fact. 15(1), 131 (2016).
Li, Q. A. et al. Ligand binding induces an ammonia channel in 2-amino-2-desoxyisochorismate (ADIC) synthase PhzE. J. Biol. Chem. 286(20), 18213–18221 (2011).
Jin, X. J. et al. iTRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis P3. Sci. Rep. 6, 27393 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Hu, H. et al. Production of trans-2,3-dihydro-3-hydroxyanthranilic acid by engineered Pseudomonas chlororaphis GP72. Appl. Microbiol. Biotechnol. 101(17), 6607–6613 (2017).
Weydert, C. J. & Cullen, J. J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 5, 51–66 (2010).
Martins, D. et al. Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 115, 9797–9802 (2018).
Blankenfeldt, W. et al. Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens. Proc. Natl. Acad. Sci. U. S. A. 101(47), 16431–16436 (2004).
Chin, A. W. T. F. et al. The Pseudomonas chlororaphis PCL1391 sigma regulator psrA represses the production of the antifungal metabolite phenazine-1-carboxamide. Mol. Plant Microbe Interact. 18(3), 244–253 (2005).
Peng, H. et al. Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microb. Cell Fact. 17(1), 117 (2018).
Huang, L. et al. Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Appl. Microbiol. Biotechnol. 89(1), 169–177 (2011).
Wang, D. et al. Differential regulation of phenazine biosynthesis by RpeA and RpeB in Pseudomonas chlororaphis 30–84. Microbiology 158(Pt 7), 1745–1757 (2012).
Whistler, C. A. & Pierson, L. S. 3rd. Repression of phenazine antibiotic production in Pseudomonas aureofaciens strain 30–84 by RpeA. J. Bacteriol. 185(13), 3718–3725 (2003).
Juminaga, D. et al. Modular engineering of L-tyrosine production in Escherichia coli. Appl. Environ. Microbiol. 78(1), 89–98 (2012).
Shtark, O., Shaposhnikov, A. I. & Kravchenko, L. V. The production of antifungal metabolites by Pseudomonas chlororaphis grown on different nutrient sources. Mikrobiologiia 72(5), 645–650 (2003).
Kerins, M. J. & Ooi, A. The roles of NRF2 in modulating cellular iron homeostasis. Antioxid. Redox Signal. 29(17), 1756–1773 (2018).
Silby, M. W. et al. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 35(4), 652–680 (2011).
Liu, H. et al. Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Curr. Microbiol. 54(4), 302–306 (2007).
Shen, X. et al. Genome sequence of Pseudomonas chlororaphis GP72, a root-colonizing biocontrol strain. J. Bacteriol. 194(5), 1269–1270 (2012).
Paulsen, I. T. et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23(7), 873–878 (2005).
Nouwens, A. S. et al. Complementing genomics with proteomics: The membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis 21(17), 3797–3809 (2000).
Lee, T. S. et al. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J. Biol. Eng. 5, 12 (2011).
Parsons, J. F. et al. Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2–79. Biochemistry 43(39), 12427–12435 (2004).
Xu, N. et al. Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis. Acta Crystallogr. D Biol. Crystallogr. 69(Pt 8), 1403–1413 (2013).
Jiang, M. & Zhang, H. Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli. Curr. Opin. Biotechnol. 42, 1–6 (2016).
Noda, S. et al. Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab. Eng. 33, 119–129 (2016).
Sengupta, S. et al. Metabolic engineering of a novel muconic acid biosynthesis pathway via 4-hydroxybenzoic acid in Escherichia coli. Appl. Environ. Microbiol. 81(23), 8037–8043 (2015).
Gosset, G. Production of aromatic compounds in bacteria. Curr. Opin. Biotechnol. 20(6), 651–658 (2009).
Kramer, M. et al. Metabolic engineering for microbial production of shikimic acid. Metab. Eng. 5(4), 277–283 (2003).
Chancey, S. T. et al. Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30–84 in the wheat rhizosphere. Appl. Environ. Microbiol. 68(7), 3308–3314 (2002).
Heeb, S., Blumer, C. & Haas, D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184(4), 1046–1056 (2002).
Slininger, P. J. & Shea-Wilbur, M. A. Liquid-culture pH, temperature, and carbon (not nitrogen) source regulate phenazine productivity of the take-all biocontrol agent Pseudomonas fluorescens 2–79. Appl. Microbiol. Biotechnol. 43(5), 794–800 (1995).
van Rij, E. T. et al. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol. Plant Microbe Interact. 17(5), 557–566 (2004).
Acknowledgements
This little article has been posted as a preprint on Research Square (www.researchsquare.com).
Funding
This work was financially supported by Focus on Research and Development Plan in Shandong Province (Nos. 2019JZZY011003, 2020CXGC010603); Shandong key project of Research & Development plan (No. 2019GSF107066); Young doctorate Cooperation Fund Project, QiLu University of Technology (Shandong Academy of Sciences) (No. BSHZ20180016); Natural Science Foundation of Shandong Province (Nos. ZR2019PC060, ZR2020QC044); Shandong Province Higher Educational Science and Technology Program (No. A18KA116); Open Project Program of State Key Laboratory of Biobased Material and Green Papermaking, Qilu University of Technology (No. KF201822). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
L.L. and W.W. conceived and designed the experiments. K.L. and W.Y. performed the experiments. Y.H., R.W., and P.L. analyzed the data. L.L. and K.L. drafted the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, K., Li, L., Yao, W. et al. Genetic engineering of Pseudomonas chlororaphis Lzh-T5 to enhance production of trans-2,3-dihydro-3-hydroxyanthranilic acid. Sci Rep 11, 16451 (2021). https://doi.org/10.1038/s41598-021-94674-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94674-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.