Abstract
Visna/maedi (VM) is a multisystemic lentivirus infection of sheep that affecting sheep industry across the globe. TMEM154 gene has been identified to be a major VM-associated host gene, nevertheless, a recent study showed that the frequency of the VM-resistant TMEM154 haplotypes was very low or absent in indigenous sheep. Thus, the present study was designed to determine other possible co-receptors associated with VM. For this purpose, DRB1 gene, which is renowned for its role in host immune response against various diseases was targeted. A total number of 151 case–control matched pairs were constructed from 2266 serologically tested sheep. A broad range of DRB1 haplotype diversity was detected by sequence-based genotyping. Moreover, a novel 2 bp deletion (del) in the DRB1 intron 1 was identified. For the final statistic, the sheep carrying VM-resistant TMEM154 diplotypes were removed and a McNemar’s test with a matched pairs experimental design was conducted. Consequently, it was identified for the first time that the 2 bp del variant is a genetic risk factor for VM (p value 0.002; chi-square 8.31; odds ratio 2.9; statistical power 0.90) in the dominant model. Thus, negative selection for 2 bp del variant could decrease VM infection risk in Turkish sheep.
Similar content being viewed by others
Introduction
Small ruminant lentiviruses (SRLVs) such as caprine arthritis encephalitis virus (CAEV) in goats and Visna/maedi virus (VMV) in sheep cause prevalent chronic infections across the globe. The infection results in multisystemic inflammation like arthritis, mastitis, lymphadonopathy, interstitial pneumonia, and meningoencephalitis. SRLV infections are mainly characterized by an insidious onset, slow disease progression, and ultimate fatality. Both viruses (CAEV and VMV) are similar in their genetic structure that paves the way for the cross-species infection transmission between sheep and goat1,2,3.
Visna/maedi (VM) has been reported in Europe4,5, North America6, South America7, Asia8, and Africa9,10. Only New Zealand and Australia are yet VM free, nonetheless, these countries are suffering from CAEV11. VM was first reported in Turkey in 198712. Regardless of sampling methods, successive reports showed that VM seroprevalence was from 2.7 to 77.9%13,14,15. In a recent serosurvey on randomly sampled seven native Turkish breeds and four crossbreds (n = 2266), it was demonstrated that the breed-level mean seroprevalence was 12.7% ranging from 2 to 83.1%. In the same study, 13 out of 16 flocks were seropositive, and the flock level prevalence was 81.3%16.
There is no effective vaccine against VM as of the present. Once the animal acquires the VM virus, it will remain as a life long reservoir of the virus due to the persistency of infection1. Several studies reported indirect production losses caused by VM through low conception rates, reduced milk production, lower birth or weaning weight of lambs given by infected ewes17. On the other side, death or premature culling of infected animals are direct production losses18. The estimated VM associated production loss was up to 20% in the USA19 and 40% in UK17.
Testing and culling VM positive ewes and their lambs, the requirement of extra facilities to keep positive sheep until culling, and restocking ewes make the infection eradication efforts much more difficult in terms of cost and labor20. Moreover, screening and culling eradication strategy for VM does not guarantee long-lasting infection-free flocks as flocks will remain open to a new infection at any time. A typical example of such case is a field trial in 1979, where thirteen VM affected commercial flocks with 17% mean seroprevalence were subjected to screening and culling program for every 6 months. After two years, at the 5, 6, and 7th screening, all flocks were found seronegative. However, at the 8th test, three VM seropositive sheep were found where the exact source of the infection remained unclear21.
The search for ways to implement effective selective breeding strategies against lethal and chronic diseases like VM has been gaining increased attention worldwide. Several candidate loci such as ovar-DRB122,23, CCR524, DPPA2/DPPA4 and SYTL325, ZNF38926, and TLR927 were reported to be associated with the VM serostatus and/or VM virus proviral load. Moreover, a case–control matched pairs experimental design revealed two haplotypes (haplotypes 1 and 4) in Exon 2 region of TMEM154 gene having a major effect according to the recessive model on reducing susceptibility to VM in North American sheep28. Subsequent studies have confirmed this association in North American29,30, German, Iranian31,32, and Turkish sheep16. The frequencies of the protective TMEM154 haplotypes, however, were reported to be significantly low or absent in some indigenous Turkish and Iranian sheep16,32. TMEM154 encodes a transmembrane protein highly expressed in B lymphocytes. There are evidence that TMEM154 variants associated with type 2 diabetes susceptibility in human33,34. Despite the effect of TMEM154 on VM occurrence has been repeatedly reported, its biological role in the host response pathway against VM is not yet known. Although TMEM154 has been identified to be a major gene regarding resistance/susceptibility to VM, there could be other possible co-receptors that affect VM occurrence. In case the possible co-receptors are detected; they could be used in selective breeding for VM in indigenous sheep breeds.
Earlier attempts by our team to investigate the association between some previously reported VM associated genes, i.e., CCR5, ZNF389, TLR9 and VM serostatus did not reveal any significant result when a case–control experimental design was implemented (unpublished data). Here we aimed to investigate the possible link between Ovar-DRB1 and VM serostatus. Ovar-DRB1 was reported to be associated with VM in two independent studies22,23. DRB1 is a MHC class II gene that encodes antigen-presenting receptor glycoproteins called histocompatibility molecules and plays a crucial role in recognizing peptides of pathogens and presenting them to the T-lymphocytes that eventually triggers host immune response. Because of its function in the immune system, DRB1 gene has been associated with various diseases in sheep as well as in other mammalians (reviewed in35).
In the present study, a case–control matched pairs experiment was carried out to investigate the possible association between Ovar-DRB1 variants and serostatus of VM in Turkish sheep. Furthermore, we have performed computational functional analysis to predict peptide–protein affinity and the possible effect of amino acid substitutions.
Results
Sequences obtained by using standard and universal M13 tagged primers showed 100% concordance. A total number of sixty-five DRB1 Exon 2 haplotypes were estimated across all breeds. 10:01, 13:01, 09:02, and 08:01 were the most prevalent haplotypes which occurred at an overall frequency greater than 0.05, i.e., 0.19, 0.12, 0.09, and 0.09, respectively. Some of the haplotypes were breed-specific, whereas some haplotypes (n = 21) found as single copy across all sample set. Regarding the haplotypes which were detected at least two times across all breeds, broad haplotype diversity was observed in Kivircik (31), Bandirma (29), Merino (20), and Imroz (13; Fig. 1; Supplemental table S1). The principal components (PC) plot for PC1 (21.8%) versus PC2 (20.3%) demonstrated that SBA crosses, Hampshire crosses, Ramlic and native Chios were slightly clustered regarding DRB1 genotypes according to breeds and the most distinct breed was Imroz (Fig. 2).
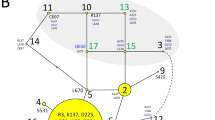
Alongside DRB1 Exon 2 haplotypes, a 2 bp deletion mutation (2 bp del) was identified at the Intron1 region of DRB1 gene, just 11 bp upstream of Exon 2 (Fig. 3). Deleted nucleotides are “CG” and the exact genomic position of the deletion mutation is 20:27300971-72 (NC_040271.1). Strong Linkage Disequilibrium (LD) was observed between the 2 bp del mutation and 13:01 haplotype (D’ value, 0.984; r-squared, 0.836), moreover, the highest LD was identified between the 2 bp del mutation and detected haplogroup 13 (haplotypes 13:01 and 13:03; D′ value, 0.984; r-squared, 0.903; Fig. 4). Haplotype 13:02 was not informative for LD analysis as it was detected as only one copy. The 2 bp del mutation was not found in Karacabey merino, Ramlic, and Hampshire crosses. The frequencies of the 2 bp del mutation in Chios, Imroz, Kivircik, Bandirma, and SBA was observed at a range of 0.03 (Imroz) to 0.42 (SBA; Table 1).
A McNemar’s test was performed over 142 matched pairs to investigate the correlation between the 2 bp del mutation or the most prevalent haplotypes (10:01, 13:01, 09:02, and 08:01) and VM serostatus (Supplemental table S2). Both recessive and dominant models were tested. In McNemar’s test, the sum of the discordant pairs (1;0 and 0;1) is expected to be higher than 25 for statistical significance. Accordingly, all of the tested variants were observed to have higher than 25 discordant pairs for the dominant model but not for the recessive model. A statistically significant correlation was observed between VM serostatus and haplotype 13:01 and the 2 bp del mutation according to the dominant model (Table 2), among these two variants, however, the most significant association was detected between the 2 bp del mutation and VM serostatus (exact p value, 0.005; chi-square, 6.89; odds ratio, 2.36; CI 95) upon computing the dominant model. Statistical power analysis was performed using real sample size (142 matched pairs) and percentage of discordant pairs (33%). Hence, the statistical power of the first analysis was calculated to be 0.87 (p < 0.05).
Alongside to strong LD between the 2 bp del and haplotype 13:01, it was identified that Histidine (H) or Tyrosine (Y) substitution to Threonine (T) at codon 61 (H/Y61T) was also linked to both 2 bp del and 1301 haplotype (Fig. 4). Among 124 ovine DRB1 haplotypes deposited in Immuno Polymorphism Database (IPD), the 61T amino acid variant is specific to 13:01, 13:02, 13:03, 05:01:01, 05:02:01, 05.03:01, 26:01, and 29:01 haplotypes of which only 13:01, 13:02, 13:03, and 26:01 were detected in the present study with the overall frequencies of 0.12, 0.003, 0.007, and 0.005, respectively. Another LD analysis was performed over the 2 bp del mutation and the haplogroup 13 (haplotypes 13:01, 13;02, and 13:03) which are carrier of 61T missense variants. Since haplotype 26:01 was detected as three copy across all breeds and neither those were not carrier of 2 bp del variant, this haplotype was excluded from this analysis. Eventually, almost a perfect LD was observed between the 2 bp del variant and the 61T amino acid carrier haplogroup 13 (D′ value, 0.985; r-squared ≥ 0.903; Fig. 4). Thus, second McNemars’s test for the haplogroup 13 with 44 discordant pairs revealed a similar association with the 2 bp del mutation (exact p value, 0.013; odds ratio, 2.14) in dominant model (Table 2).
Our case–control matched pairs panel was also available for TMEM154 genotypes which were identified to have a major effect for the recessive model on VM resistance/susceptibility16,28,29,30,31,32. Finally, to test the relative protection of the wild type DRB1 genotype compared to the 2 bp del variant in the absence of the protective effect of TMEM154, these matched pairs were sorted and analyzed again. Briefly, if at least one member of a pair is a carrier of protective TMEM154 diplotypes (1;1, 1;4, or 4;4), these pairs were removed from the data set, thus, 92 matched pairs lacking the protective TMEM154 diplotypes remained (Supplemental table S3). Consequently, the third McNemar’s test for correlated proportion was performed on this data set with 39 discordant pairs (1;0 and 0;1). It was determined that 2 bp del mutation was still significantly associated with increased susceptibility, and the wild type ones were associated with relative resistance to VM despite lacking the protective TMEM154 diplotypes (exact p value, 0.002; chi-square, 8.31; odds ratio, 2.9; CI 95). The statistical power of this analyzis was calculated to be 0.90 (odds ratio, 2.9; CI 95; p < 0.05). According to our results, the 2 bp del mutation in DRB1 Intron1 was identified as a genetic risk factor in dominant model, i.e., having one or two copies of 2 bp del mutation was found to increase the risk of contracting the VM virus by 2.9 fold (Table 2).
In silico peptide–protein docking analysis was performed to predict the docking affinity between VM virus Gag protein antigenic epitope and DRB1 high-frequency haplotypes i.e.13:01, 10:01, 08:01, and 09:02. The prediction result revealed docking scores were variable among these four haplotypes and both of the two tools (HDOCK and HEPDOCK) computed relatively lower docking score for VM associated haplotype 13:01 when compared to other common haplotypes (Table 3). Additionally, functional analysis of point mutation effect was predicted for H/Y61T substitutions using two popular web applications. Accordingly, PROVEAN predicted the H61T and Y61T substitutions have “deleterious” effect while, PANTHER predicted the same substitutions to exhibit “probably benign” effect (Table 4).
Discussion
As in many other mammalians, there are several identified ovine DRB1 gene variants. According to IPD records, 124 ovine DRB1 alleles or subtypes have been deposited so far. In the present study, a broad genetic diversity in Exon 2 region of DRB1 gene was identified in Turkish sheep. There were 44 different haplotypes detected in at least two times (Supplemental table S1). The highest relative allelic diversity (the number of the detected different alleles over total alleles in each breeds) was in Chios breed (0.625) and the lowest in Kivircik breed (0.176).
Exon 2 region of the ovar DRB1 gene has gained great attention of researchers having a broad genetic diversity and its key role in immune defense. Previously, the association between various DRB1 alleles and different diseases such as Cystic Echinococcosis36, faecal egg count of gastrointestinal nematodes37,38 have been reported. The association between DRB1 gene and resistance/susceptibility to VM have also been reported in two different studies. In the first study, it was found that DRB1 haplotypes 04:03 and 07:12 were significantly associated with lower provirus levels of the ovine progressive pneumonia22, which is the counterpart of VM in North America, and in the second study, it was reported that the haplotype 03:25 was associated with the susceptibility to VM23. However, these reported haplotypes were not detected in the present study.
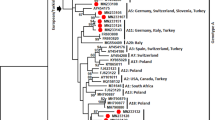
According to present findings, strong LD was detected between 2 bp del variant and 61T amino acid substitution which was found in 13:01 and 13:03 haplotypes in our genotype panel. To further investigate, fifty Ovar-DRB1 sequences consisting of 2 bp del variant in intron 1 were obtained from GenBank (Accession numbers: MG000511.1, MG000512.1, MG000515.1, MG000516.1, MG000518.1, MG000538.1 to 552.1, and MG000555.1 to 586.1). All the sequences belonged to the Djallonke and Sahelian native sheep breeds of Ghana in West Africa. These sequences were searched using BLAST on IPD server, and the results revealed that all of these sequences were compatible with the haplotype 13:02 (D′ value, 1; r-squared, 1). Archaeological evidence and retrovirus integration analyses suggest that selection for desired traits common to modern sheep first began in Fertile Crescent including the Anatolia region of Turkey, and spread to Africa, Europa, and other parts of Asia39. All the native breeds in the present study were carriers of the 2 bp del mutation, but improved breeds by backcrossing (Karacabey merino and Ramlic) were not. Other two research flocks, Bandirma which was bred from native Kivircik breed and SBA crosses, were also carriers of this mutation. When the GenBank records and our findings evaluated together; it can be inferred that having been found in African and Turkish native breeds, 2 bp deletion is an ancient mutation and perfectly linked to both haplogroup 13 (13:01, 13:02, and 13:03) and 61T amino acid variation. Furthermore, it could be speculated that 61T amino acid substitution, either alone or together with other codons, may be the causative variant for increased susceptibility against VM infection. However, case control studies are required with other 61T carrier haplotypes i.e. 05:01:01, 05:02:01, 05.03:01, 26:01, and 29:01 to strengthen this hypothesis. Furthermore, the LD status between 2 bp del and 61T amino acid variant on these haplotypes remained ambiguous. Despite these limitations, it was clearly demonstrated that 2 bp del mutation in the intron 1 of the ovine DRB1 is a strong predictor for the 61T amino acid variant in haplogroup 13, and significantly associated with increased susceptibility against VM.
In a recent study, the VM resistant TMEM154 haplotypes in Turkish native sheep breeds were detected either at a very low frequency or complately absent (0 to 0.12) when compared to the improved breeds by backcrossing16. A similar observation was made in Iranian native sheep breeds32. Hence, selective breeding regarding protective TMEM154 genotypes might be a long lasting process to improve herd level genetic resistance to VM for native breeds. Alternatively, introgression or gene editing technology, i.e., CRISPR might be required for the native breeds lacking resistant TMEM154 haplotypes. Otherwise, 2 bp del mutation and linked H/Y61T amino acid substitution was found a range of 3–25% indicating that negative selection for 2 bp del mutation could provide a chance for relatively rapid genetic improvement against VM disease for some native sheep.
From the first matched pairs panel, 21 ewes were carriers of susceptible DRB1 2 bp del variant and protective TMEM154 diplotypes (1;1, 1;4, or 4;4) at the same time. According to the previous reports, TMEM154 protective diplotypes provide approximately threefold protection to VM, thus, it is expected that only 1/4 of these 21 ewes could be VM positive. Ten of these ewes, however, were VM positive which might indicate the 2 bp del variants potentially limits the protective effect of the TMEM154. Thus, further genetic improvement could be achieved using combination of these two genetic markers (DRB1 and TMEM154) for selective breeding of both native and improved sheep breeds.
Genotyping of MHC II locus, including DRB genes is highly problematic due to the excessive variation in a relatively short exon often causes sequence reading errors. Therefore, bidirectional sequencing is needed to reduce bias. In fact, bidirectional sequencing does not guarantee error-free genotyping, therefore, it is highly recommended to clone sequences for increased precision while genotyping. However, detection of 2 bp del variation could be much easier and cost effective with availability of genotyping techniques such as Allele Specific PCR (AS-PCR), Restriction Fragment Length Polymorphism (RFLP), Single Strand Conformationel Polymorphism (SSCP), or Real-Time PCR. Besides, genotyping of 2 bp del variant using such techniques might result in much fewer genotyping errors when compared to the exon sequencing.
Identifying peptides of antigen is a key step in the development of therapeutic options for many chronic infectious diseases40. Predicting viral epitope especially is cost effective and time saving41. Despite bioinformatics prediction does not mimics experimental results, predicting epitopes plays important roles. The identified Gag protein antigenic epitope was relatively weakly bind a 2 bp del associated DRB1 receptor allele i.e. 13:01 compared to the other high-frequency alleles i.e.10:01, 08:01 and 09:02. The variation in binding affinity and peptide core could be due to the highly polymorphic nature of DRB1 especially the specific binding cores42. This finding possibly strengthen the susceptibility feature of 13:01 because of the poorly docking affinity between the antigenic epitope of VM virus and this haplotype which potentially interferes the immune response upon infection.
As observed in this study, H/Y61T substitution was strongly associated with 13 haplogroup. We hypothesized that the variant may maneuver the receptor core affinity to the viral antigenic epitope. This assumption is supported by the proclivity of peptides to bind to DRB1 of a particular allele in human43. Similarly, variants of peptides could define ligand specific binding capacity44 which in turn is defined by the docking energy score. Further to that, according to PROVEAN web server result, the new variant (T) was predicted to have deleterious effect than the commonly observed variants at position 61. The results obtained from PANTHER and PROVEAN servers were slightly inconsistent. This could be due to the assumptions that the databases are based on. PROVEAN takes the 75% sequence homology modeling to predict the effect of amino acid substitutions while PANTHER is based on the evolutionary history of a domain assuming a preserved region has functional importance. Additionally, PANTHER predicts the function of a query against the already existing 100 species in the databases which does not include Ovis aries. The sum of the results in terms of docking affinity and predictive effect of point mutation could give us a clue on how different alleles and variants that are harbored in the haplotypes affect the cellular immune reaction in fighting against the viral invasion.
In conclusion, this is a pioneering study in the identification of the association between the 2 bp del variation in ovine DRB1 intron 1 and VM serostatus in the presence and/or absence of the protective effect of the major gene TMEM154. The protective haplotypes of TMEM154 were previously detected at a high frequency in improved breeds such as Karacabey merino and Ramlic, and for native breeds they were in very low frequencies or absent. Conversely, 2 bp del variant of the DRB1 gene having a strong association with increased susceptibility to VM was found in many native Turkish breeds. Thus, considering the prevalence of VM in some native breeds, it is highly reasonable to take the DRB1 2 bp del variant into account for selective breeding to obtain infection-free sheep flocks.
Methods
Animals
In this study, three indigenous Turkish breeds (Chios, Imroz, and Kivircik), two improved breeds (Karacabey Merino and Ramlic), and three composite breeds (Blackhead merino crosses-SBA, Hampshire crosses, and Bandirma) were considered. Chios, Imroz, and Kivircik are ancient breeds that are well-adapted to their environment for thousands of years. Karacabey merino was improved by backcrossing in the 1940s in Sheep Breeding and Research Institute (SRI), whereas Ramlic was improved in the 1960s by the Turkish Ministry of Agriculture and Forestry. Both breeds have been closed for backcrossing for more than 30 years. The composite breeds: SBA, Hampshire, and Bandirma have been reared as research flocks at SRI. Ewes for the study were selected according to their serostatus, age, breed type, and flock from the 2017 serosurvey cohort of 2266 ewes of eleven flocks at six different locations16. To allow sufficient VM virus exposure and subsequent seroconversion, only the ewes that were two years or older ages were included (ages ranging from two to eight). Thus, a total number of 302 ewes (151 cases and 151 controls) were matched. Haplotype phasing and LD analysis were conducted over these matched pairs. All methods were performed in accordance with the relevant guidelines and regulations.
Serological analysis
To determine the VM virus-specific antibody titer in the serum samples, an Enzyme-Linked Immunosorbent Assay (ELISA) was carried out. Briefly, serums were separated from fresh whole blood samples by centrifugation and were tested with Idexx CAEV/VMV total Ab ELISA commercial kit following the manufacturer’s manual. At the final step, ELISA plates were read at 450 nm wavelength by an ELISA plate reader. The sensitivity and specificity of the used ELISA kit have reported being 84.3% and 99.6%, respectively45.
Genetic analysis
Genomic DNA was extracted from peripheral whole blood with EDTA using commercial spin-column DNA extraction kits according to the manufacturer’s instructions. Polymerase chain reaction (PCR) was carried out to amplify Exon 2 region of Ovar-DRB1 gene using previously designed primers (DRB1_330_F: ATTAGCCTCYCCCCAGGAGKC and DRB1_329_R: CACCCCCGCGCTCACCTCGCCGC)46. Amplified products overlap the last 54 bp of DRB1 Intron1 and 270 bp of the full sequence of Exon 2. Standard Sanger sequencing analysis was performed as follows: pre-purification of PCR products by Exo-SAP incubation followed by chain termination reaction with BigDye™ Terminator v3. 1 kit, post-purification with ethanol precipitation protocol, and capillary electrophoresis on AB 3500 genetic analyzer were performed sequentially. All samples were sequenced bidirectionally. Approximately 1/4 (28%) from all samples were selected randomly and amplified separately using the same primers with universal M13 primer tags (M13_F: GTAAAACGACGGCCAGT and M13_R: AACAGCTATGACCATG) and sequenced bidirectionally to validate the obtained sequences and check primers binding sites.
Statistical analysis
Chromatograms were visualized using FinchTV v1.4.0 software (Geospiza, Inc) and aligned by MEGA v6.0 software47. DRB1 Exon 2 haplotypes were estimated using Phase v2.1 algorithm48 and identified by Basic Local Alignment Search Tool (BLAST) in both National Center for Biotechnology (NCBI; https://www.ncbi.nlm.nih.gov/) and Immuno Polymorphism Database (IPD; https://www.ebi.ac.uk/ipd/). LD analysis was performed using Haploview v4.2 2049.
Host genotype and disease resistance/susceptibility association study requires the control of confounding factors such as variations in exposure intensity, exposure duration and other environmental factors such as climate, herd management, herd density, access to the pasture. Furthermore, it is required to increase statistical power to reduce false positive results potentially caused by the population stratification phenomenon. Therefore, association analysis was performed according to case–control matched pairs experimental design. A total of 151 matched pairs (151 cases and 151 controls) were constructed according to age, breed type, and flock. Briefly, a seropositive ewe was randomly matched with a seronegative one from the same flock, same breed type, and same age group. This matched pairs panel was used for haplotype phasing and LD analiysis. Due to limited animal numbers in SBA (6) and Hampshire crosses (6), Ramlic (2), and native Chios (4), these breeds were ignored, and statistical analysis was performed on the remaining 142 matched pairs (Table 5). The age information was not available for a small proportion of matched pairs panel (4.2%). Thus, these individuals were matched according to breed type and flock. The pairs were assigned to be (1;1), (1;0), (0;1), and (0;0) i.e. both case and control are carriers of genetic risk/protective factors (1;1), the case is the carrier of genetic risk/protective factors, but control is not (1;0), the case is not a carrier of genetic risk/protective factors, but control is (0;1), and both case and control are not carriers of genetic risk/protective factors (0;0). In this experimental design, the higher rate of the discordant pairs (1;0 and 0;1) provides higher statistical power. A McNemar’s test50 for correlated proportions was performed using these matched pairs. To determine the statistical power of the experimental design, a power analysis was conducted using the G*Power software51.
In silico analysis
The binding affinity of the DRB core to the peptide is a potential factor in presenting antigen to immune cells52. Identifying the viral epitope is the first step to calculate the affinity of the given antigenic peptide to the DRB cores. Gag protein epitope of VM virus was extracted from Immune Epitope Database and Analysis Resource (IEDB, http://www.immuneepitope.org). Gag protein epitope was used for further ligand–protein affinity analysis using HEPDOCK and HDOCK web servers53. Besides, the effect of the H/Y61T substitution which is specific to 2 bp del linked haplogroup 13 (13:01, 13:02, and 13:03) was predicted using PROVEAN (http://provean.jcvi.org/) and PANTHER (http://www.pantherdb.org) web applications.
Ethics declarations
All animal procedures in the study were reviewed and approved by the local ethics committee of Sheep Breeding and Research Institute (Approval No. 1282412), and the authors complied with the ARRIVE guidelines.
References
Pépin, M., Vitu, C., Russo, P., Mornex, J.-F. & Peterhans, E. Maedi–Visna virus infection in sheep: A review. Vet. Res. 29, 341–367 (1998).
Shah, C. et al. Direct evidence for natural transmission of small-ruminant lentiviruses of subtype A4 from goats to sheep and vice versa. J. Virol. 78, 7518–7522 (2004).
Reina, R. et al. Prevention strategies against small ruminant lentiviruses: An update. Vet. J. 182, 31–37 (2009).
Hüttner, K., Heyne, H. & Heim, D. Impact of flock segregation according to the maedi–Visna status on reproduction and lamb rearing—A field study in Mecklenburg–Vorpommern. Berl. Munch. Tierarztl. Wochenschr. 130, 490–493 (2017).
Ogden, N., Davies, P. & Lovatt, F. Dealing with maedi–Visna in UK sheep flocks. Practice 41, 321–328 (2019).
Heinrichs, R., Wilkins, W., Schroeder, G. & Campbell, J. Prevalence of maedi–Visna in Saskatchewan sheep. Can. Vet. J. 58, 183–186 (2017).
Alves, S. M. et al. Seroepidemiological study of maedi–visna in sheep in Ceara, Rio Grande do Norte, Paraíba, and Sergipe States Semina. Ciênc. Agrár. 39, 2017–2028 (2018).
Oguma, K. et al. Isolation of Maedi/Visna virus from a sheep in Japan. J. Vet. Med. Sci. 76, 211–218 (2014).
Ayelet, G., Roger, F., Tibbo, M. & Tembely, S. Survey of maedi–Visna (MV) in Ethiopian highland sheep. Vet. J. 161, 208–210 (2001).
Tabet, E., Tlaige, R., El Hage, J. & Abi-Rizk, A. The occurrence of maedi–Visna virus in Lebanon. Rev. Sci. Tech. 36, 899–903 (2017).
Gomez-Lucia, E., BaVrquero, N. & Domenech, A. Maedi–Visna virus: Current perspectives. Vet. Med. (Auckl). 21, 11–21 (2018).
Girgin, H., Aydın, N., Yonguç, A. D., Aksoy, E. & Çorak, R. Ve şimdi koyunların Viral Maedi–Visnası Türkiye’de. Etlik. Vet. Mikrob. Derg. 9, 20 (1987).
Burgu, İ et al. Türkiye’de Visna–Maedi enfeksiyonunun serolojik olarak araştırılması. A Ü Vet. Fak. Derg. 37, 538–553 (1990).
Yavru, S., Şimşek, A., Bulut, O. & Kale, M. Serological investigation of Maedi–Visna virus infection in sheep in Konya region. Eurasian J. Vet. Sci. 28, 142–148 (2012).
Muz, D. et al. First molecular characterization of visna/Maedi viruses from naturally infected sheep in Turkey. Arch. Virol. 158, 559–570 (2013).
Yaman, Y. et al. Association of TMEM154 variants with visna/Maedi virus infection in Turkish sheep. Small Rum. Res. 177, 61–67 (2019).
Benavides, J. et al. Impact of maedi–visna in intensively managed dairy sheep. Vet. J. 197, 607–612 (2013).
Peterhans, E. et al. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 35, 257–274 (2004).
Keen, J. E., Hungerford, L. L., Littledike, E. T., Wittum, T. E. & Kwang, J. Effect of ewe ovine lentivirus infection on ewe and lamb productivity. Prev. Vet. Med. 30, 155–169 (1997).
Fisher, J. W. & Menzies, P. I. Cost of a Maedi–Visna flock certification program and the changes in productivity and economic return. Sheep Goat Res. J. 20, 17–24 (2005).
Houwers, D. J., Schaake, J. & de Boer, G. F. Maedi–visna control in sheep II. Half-yearly serological testing with culling of positive ewes and progeny. Vet. Microbiol. 9, 445–451 (1984).
Herrmann-Hoesing, L., White, S. N., Mousel, M. R., Lewis, G. S. & Knowles, D. P. Ovine progressive pneumonia provirus levels associate with breed and Ovar-DRB1. Immunogenetics 60, 749–758 (2008).
Larruskain, A. et al. MHC class II DRB1 gene polymorphism in the pathogenesis of Maedi–Visna and pulmonary adenocarcinoma viral diseases in sheep. Immunogenetics 62, 75–83 (2010).
White, S. N., Mousel, M. R., Reynolds, J. O., Lewis, G. S. & Herrmann-Hoesing, L. M. Common promoter deletion is associated with 3.9-fold differential transcription of ovine CCR5 and reduced proviral level of ovine progressive pneumonia virus. Anim. Genet. 40, 583–589 (2009).
White, S. N. et al. Genome-wide association identifies multiple genomic regions associated with susceptibility to and control of ovine lentivirus. PLoS ONE 7, e47829 (2012).
White, S. N., Mousel, M. R., Reynolds, J. O., Herrmann-Hoesing, L. M. & Knowles, D. P. Deletion variant near ZNF389 is associated with control of ovine lentivirus in multiple sheep flocks. Anim. Genet. 45, 297–300 (2014).
Sarafidou, T. et al. Toll Like Receptor 9 (TLR9) polymorphism G520R in sheep is associated with seropositivity for small ruminant lentivirus. PLoS ONE 8, e63901 (2013).
Heaton, M. P. et al. Reduced lentivirus susceptibility in sheep with TMEM154 mutations. PLoS Genet. 8, 467 (2012).
Leymaster, K. A., Chitko-McKown, C. G., Clawson, M. L., Harhay, G. P. & Heaton, M. P. Effects of TMEM154 haplotypes 1 and 3 on susceptibility to ovine progressive pneumonia virus following natural exposure in sheep. J. Anim. Sci. 91, 5114–5121 (2013).
Leymaster, K. A., Chitko-Mckown, C. G. & Heaton, M. P. Incidence of infection in 39-month-old ewes with TMEM154 diplotypes “1 1,” “1 3,” and “3 3” after natural exposure to ovine progressive pneumonia virus. J. Anim. Sci. 93, 41–45 (2015).
Molaee, V., Eltanany, M. & Lühken, G. First survey on association of TMEM154 and CCR5 variants with serological maedi–visna status of sheep in German flocks. Vet. Res. 49, 36 (2018).
Molaee, V., Otarod, V., Abdollahi, D. & Lühken, G. Lentivirus susceptibility in Iranian and German sheep assessed by determination of TMEM154 E35K. Animals 9, 685 (2019).
Mahajan, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 46, 234–244 (2014).
Harder, M. et al. The type 2 diabetes risk allele of TMEM154-rs6813195 associates with decreased beta cell function in a study of 6,486 Danes. PLoS ONE 10, 0120890 (2015).
Dukkipati, V. S. R., Blair, H. T., Garric, D. J. & Murray, A. Ovar-Mhc-ovine major histocompatibility complex: Role in genetic resistance to diseases. N. Z. Vet. J. 54, 153–160 (2006).
Shen, H., Han, G., Jia, B., Jiang, S. & Du, Y. MHC-DRB1/DQB1 Gene polymorphism and its association with resistance/susceptibility to Cystic Echinococcosis in Chinese Merino Sheep. J. Parasitol. Res. 2014, 272601 (2014).
Charon, K. M., Moskwa, B., Rutkowski, R., Gruszczyńska, J. & Świderek, W. Microsatellite polymorphism in DRB1 gene (MHC class II) and its relation to nematode faecal egg count in Polish heath sheep. J. Anim. Feed. Sci. 11, 47–58 (2002).
Sayers, G. et al. Major histocompatibility complex DRB1 gene: Its role in nematode resistance in Suffolk and Texel sheep breeds. Parasitology 131, 403–409 (2005).
Chessa, B. et al. Revealing the history of sheep domestication using retrovirus integrations. Science 324, 532–536 (2009).
Vilas-Boas, L. C. P. et al. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 76, 3525–3542 (2019).
Sakib, M. S., Islam, M. R., Hasan, A. K. & Nabi, A. H. Prediction of epitope-based peptides for the utility of vaccine development from fusion and glycoprotein of nipah virus using in silico approach. Adv. Bioinf. 2014, 402492 (2014).
Vogt, A. B. et al. Ligand motifs of HLA-DRB5*0101 and DRB1*1501 molecules delineated from self-peptides. J. Immunol. 153, 1665–1673 (1994).
Jiang, W. & Eric, T. B. High-throughput engineering and analysis of peptide binding to class II MHC. Proc. Natl. Acad. Sci. U S A. 107, 13258–13263 (2010).
Malcherek, G. et al. Analysis of allele-specific contact sites of natural HLA-DR17 ligands. J. Immunol. 153, 1141–1149 (1994).
Michiels, R. et al. Comparative analysis of different serological and molecular tests for the detection of small ruminant lentiviruses (SRLVs) in Belgian sheep and goats. Viruses 10, 696 (2018).
Ballingall, K. T. & Tassi, R. Sequence-based genotyping of the sheep MHC class II DRB1 locus. Immunogenetics 62, 31–39 (2010).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Stephens, M. & Donnelly, P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 (2003).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 15, 263–265 (2005).
McNemar, Q. Note on the sampling error of the difference between correlated proportions. Psychometrika 12, 153–157 (1947).
Faul, F., Erdfelder, E., Buchne, A. & Lang, A. G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods. 41, 1149–1160 (2009).
Reynisson, B. et al. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J. Proteome Res. 19, 2304–2315 (2020).
Yan, Y., Huanyu, T., Jiahua, H. & Sheng-You, H. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15, 1829–1852 (2020).
Acknowledgements
We thank Veterinarian Ahmet İPÇİ and Veterinary technician Süleyman ÖZDEMİR for their important helps to field coordination and sampling. We dedicate this paper to the memory of our colleague Prof. Dr. Kemal Özdem ÖZTABAK.
Funding
Funding for this research was provided by the Republic of Turkey Ministry of Agriculture and Forestry, General Directorate of Agricultural Research and Policies (TAGEM) (Project No. TAGEM/HAYSÜD/15/A01/P02/02-02).
Author information
Authors and Affiliations
Contributions
Y.Y. conceived and designed the study, conducted the statistical analysis, Y.Ö and V.B performed quality controls of genomic data and conducted haplotype construction, R.A and M.K conducted laboratory experiments, E.Y.T performed in silico analiysis, and E.Y.T and C.Ü. reviewed the manuscript and assisted in data interpretation. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yaman, Y., Bay, V., Aymaz, R. et al. A novel 2 bp deletion variant in Ovine-DRB1 gene is associated with increased Visna/maedi susceptibility in Turkish sheep. Sci Rep 11, 14435 (2021). https://doi.org/10.1038/s41598-021-93864-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-93864-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.